- ImageCLEF 2025
- LifeCLEF 2025
- ImageCLEF 2024
- LifeCLEF 2024
- ImageCLEF 2023
- LifeCLEF 2023
- ImageCLEF 2022
- LifeCLEF2022
- ImageCLEF 2021
- LifeCLEF 2021
- ImageCLEF 2020
- LifeCLEF 2020
- ImageCLEF 2019
- LifeCLEF 2019
- ImageCLEF 2018
- LifeCLEF 2018
- ImageCLEF 2017
- LifeCLEF2017
- ImageCLEF 2016
- LifeCLEF 2016
- ImageCLEF 2015
- LifeCLEF 2015
- ImageCLEF 2014
- LifeCLEF 2014
- ImageCLEF 2013
- ImageCLEF 2012
- ImageCLEF 2011
- ImageCLEF 2010
- ImageCLEF 2009
- ImageCLEF 2008
- ImageCLEF 2007
- ImageCLEF 2006
- ImageCLEF 2005
- ImageCLEF 2004
- ImageCLEF 2003
- Publications
- Old resources
You are here
ImageCLEFmed GAN
Welcome to the 3rd edition of the GANs task!
Motivation
Controlling the Quality of Synthetic Medical Images created via GANs
AI systems for medical tasks like predicting, detecting and classifying diseases rely on the availability of large and diverse datasets for training. High-quality data allows these models to learn complex patterns and improve their accuracy and reliability. However, getting access to real medical data is not easy because of privacy concerns. Patients are usually willing to share their medical information only for their own treatment and not for research. This makes it hard to gather enough data to train AI models effectively, slowing down progress in developing better tools for healthcare.
One way to solve this problem is by creating synthetic data—artificial data that looks like real medical data but doesn’t come from actual patients. Generative models, like GANs (Generative Adversarial Networks), can be used to create these datasets. Synthetic data can help researchers build and test AI systems without needing to rely on real patient data, which protects privacy and makes it easier to collect the variety of information needed for training.
But there’s an important challenge with synthetic data: it must not include hidden details, or “fingerprints” from the real data it was trained on. If synthetic data can somehow be traced back to the original patient data, it could risk exposing private information. Ensuring synthetic data are completely free from such “fingerprints” is critical. It’s the only way to guarantee patient privacy while using synthetic data to push forward advancements in AI-powered healthcare.
Lessons learned:
-
In the first and second editions of this task, held at ImageCLEF 2023 and 2024, various generative models were analyzed within the framework of the first subtask to investigate whether synthetic images contained "fingerprints" of the real medical data used during training. The results demonstrated that generative models do retain and imprint features from their training data, raising important security and privacy concerns. These findings underscore the need for robust techniques to detect and mitigate such imprints to ensure that synthetic images protect patient privacy while maintaining their utility for research and development.
-
In the 2nd edition of the task, it was confirmed that generative models leave unique "fingerprints" on the synthetic images they produce. By analyzing images generated from various models, distinct patterns and features were identified that allowed the attribution of synthetic images to their respective generative models.
News
Both train and test datasets have been released and can be found here.
Task Description
We will continue to investigate the hypothesis that generative models generate synthetic medical images that retain "fingerprints" from the real images used during their training. These fingerprints raise important security and privacy concerns, particularly in the context of personal medical image data being used to create artificial images for various real-life applications.
The task is divided into two subtasks, both focusing on detecting and analyzing these "fingerprints" within synthetic biomedical image data to determine which real images contributed to the training process:
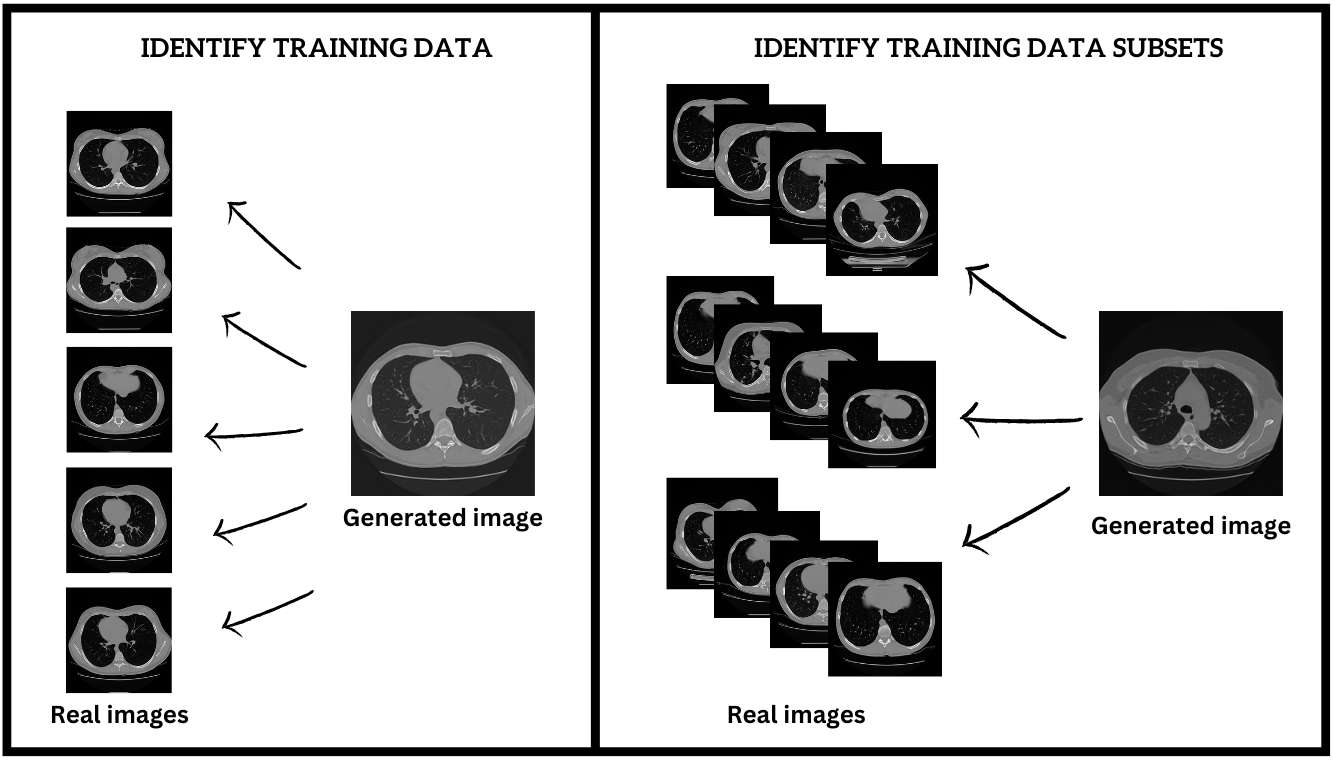
Subtask 1: Detect Training Data Usage
In this subtask, participants will analyze synthetic biomedical images to determine whether specific real images were used in the training process of generative models. For each real image in the test set, participants must label it as either used (1) or not used (0) for generating the given synthetic images. This task focuses on detecting the presence of training data "fingerprints" within synthetic outputs.
Subtask 2: Identify Training Data Subsets
In this subtask, participants will link each synthetic biomedical image to the specific subset of real data used during its generation. The goal is to identify the particular dataset of real images that contributed to the training of the generative model responsible for creating each synthetic image. This requires a more detailed attribution of synthetic images to their corresponding training subsets.
Both subtasks aim to advance our understanding of how generative models utilize training data, ensuring that synthetic data generation respects privacy and mitigates potential risks associated with patient confidentiality.
Data
The benchmarking dataset includes both real and synthetic biomedical images. The real images consist of axial slices of 3D CT scans from approximately 8,000 lung tuberculosis patients. These slices vary in appearance: some may look relatively “normal,” while others exhibit distinct lung lesions, including severe cases. The real images are stored in 8-bit per pixel PNG format, with dimensions of 256x256 pixels, providing a standardized resolution for analysis.
The synthetic images, also sized at 256x256 pixels, have been generated using various generative models, including Generative Adversarial Networks (GANs) and Diffusion Neural Networks. By providing both real and synthetic datasets, this task enables participants to analyze and compare the characteristics of synthetic images with their real counterparts, investigating potential "fingerprints" and patterns related to the training process.
Datasets are available here
Subtask 1: Detect Training Data Usage
TRAINING DATASET consists of 3 folders:
- "generated" – contains 5,000 synthetic images generated using a Generative Adversarial Network (GAN).
- "real_used" – contains 100 real images that were used to train the GAN to produce the synthetic images in the "generated" folder.
- "real_not_used" – contains 100 real images that were not used in training the GAN.
TEST DATASET consists of 2 folders:
- "generated" – This folder contains 2,000 additional synthetic images. These images were generated using the same GAN model trained under the same conditions as those used to create the synthetic images in the training dataset.
- "real_unknown" – This folder contains a mix of 500 real images. Some of these images were used in training the generative model, while others were not
Each image in the test dataset will be assigned a label. Submission files must follow the guidelines outlined in the "Submission Instructions" section.
Subtask 2: Identify Training Data Subsets
The TRAINING DATASET consists of two main folders:
- "generated" – contains 5 subfolders, each holding synthetic images. Each subset was generated using a different training dataset for the generative model.
- "real" – contains 5 subfolders, each corresponding to a specific training dataset used to train the generative model. The real images in each subfolder were used to generate the synthetic images in the corresponding "generated" subfolder.
The mapping between the real and generated images is as follows:
- Folder "t1" (real images) → Used to generate synthetic images in "gen_t1"
- Folder "t2" (real images) → Used to generate synthetic images in "gen_t2"
- Folder "t3" (real images) → Used to generate synthetic images in "gen_t3"
- Folder "t4" (real images) → Used to generate synthetic images in "gen_t4"
- Folder "t5" (real images) → Used to generate synthetic images in "gen_t5"
The TEST DATASET contains 25,000 generated images, each derived from a real subgroup of images in the training dataset. Each image will be assigned a label consistent with those used in the training dataset.
Submission files must follow the guidelines outlined in the "Submission Instructions" section
Evaluation methodology
TBD
For assessing the performance of Task 1, Cohen Kappa Score will be used.Accuracy is the official evaluation metric for Task 2.
Participant registration
Please refer to the general ImageCLEF registration instructions
Preliminary Schedule
TBA
Submission Instructions
Subtask 1: Detect Training Data Usage
- The submission must include a run file named exactly: run.csv.
- This file must be zipped; the zip file can have any name.
- The run.csv file should contain two columns in the following format:
, where 1 indicates the image was used for training and 0 indicates the image was not used for training.
Example Submission Structure:
• Method1.zip
├── run.csv (this file will be compared with the ground truth)
├── real_unknown_1.png 1
├── real_unknown_2.png 0
├── ...
├── real_unknown_500.png 1Subtask 2: Identify Training Data Subsets
- Each synthetic image must be labeled based on the real image subgroup it was generated from, using one of the following labels: [1, 2, 3, 4, 5].
- The run file must be named exactly: run.csv
- It must contain two columns formatted as:
- The run.csv file must be zipped; the zip file can have any name.
Example Submission Structure:
• Method1.zip
├── run.csv (this file will be compared with the ground truth)
├── gen_unknown_00001.png 1
├── gen_unknown_00002.png 2
├── gen_unknown_00003.png 3
├── ...
├── gen_unknown_25000.png 3Constraints to follow:
- Each test set image ID must appear exactly once in the submitted file.
- The image names must be preserved exactly as they appear in the dataset.
- Do not include any headers in the file.
Results
TBA
CEUR Working Notes
For detailed instructions, please refer to this PDF file. A summary of the most important points:
- All participating teams with at least one graded submission, regardless of the score, should submit a CEUR working notes paper.
- Teams who participated in both tasks should generally submit only one report
Citations
TBA
Contact
Organizers:
- Alexandra Andrei, alexandra.andrei(at)upb.ro, National University of Science and Technology POLITEHNICA Bucharest, Romania
- Ahmedkhan Radzhabov, National Academy of Science of Belarus, Minsk, Belarus
- Yuri Prokopchuk, National Academy of Science of Belarus, Minsk, Belarus
- Liviu-Daniel Ștefan, National University of Science and Technology POLITEHNICA Bucharest, Romania.
- Mihai Gabriel Constantin, National University of Science and Technology POLITEHNICA Bucharest, Romania.
- Mihai Dogariu, National University of Science and Technology POLITEHNICA Bucharest, Romania.
- Vassili Kovalev, vassili.kovalev(at)gmail.com, Belarusian Academy of Sciences, Minsk, Belarus
- Bogdan Ionescu , bogdan.ionescu(at)upb.ro, National University of Science and Technology POLITEHNICA Bucharest, Romania
- Henning Müller , henning.mueller(at)hevs.ch, University of Applied Sciences Western Switzerland, Sierre, Switzerland
Acknowledgments
| Attachment | Size |
|---|---|
| 260.93 KB |
