- ImageCLEF 2025
- LifeCLEF 2025
- ImageCLEF 2024
- LifeCLEF 2024
- ImageCLEF 2023
- LifeCLEF 2023
- ImageCLEF 2022
- LifeCLEF2022
- ImageCLEF 2021
- LifeCLEF 2021
- ImageCLEF 2020
- LifeCLEF 2020
- ImageCLEF 2019
- LifeCLEF 2019
- ImageCLEF 2018
- LifeCLEF 2018
- ImageCLEF 2017
- LifeCLEF2017
- ImageCLEF 2016
- LifeCLEF 2016
- ImageCLEF 2015
- LifeCLEF 2015
- ImageCLEF 2014
- LifeCLEF 2014
- ImageCLEF 2013
- ImageCLEF 2012
- ImageCLEF 2011
- ImageCLEF 2010
- ImageCLEF 2009
- ImageCLEF 2008
- ImageCLEF 2007
- ImageCLEF 2006
- ImageCLEF 2005
- ImageCLEF 2004
- ImageCLEF 2003
- Publications
- Old resources
You are here
ImageCLEFmed: The Medical Task 2016
<--
News
- 26.04.2016:The test set has been updated. The following changes were made:
- Figure separation: image "1471-244X-7-25-4" was removed
- Multi-label classification: image "1471-244X-7-25-4" was removed
- Subfigure classification: images "1471-244X-7-25-4-1", "1471-244X-7-25-4-2", "1471-244X-7-25-4-3", "1471-244X-7-25-4-4", "1471-244X-7-25-4-5", "1471-244X-7-25-4-6", "1471-244X-7-25-4-7", "1471-244X-7-25-4-8", "1471-244X-7-25-4-9", "1471-244X-7-25-4-10", "1471-244X-7-25-4-11", "1471-244X-7-25-4-12", "1471-244X-7-25-4-13", "1471-244X-7-25-4-14", "1471-244X-7-25-4-15", "1471-244X-7-25-4-16", "1471-244X-7-25-4-17", "1471-244X-7-25-4-18", "1471-244X-7-25-4-19", "1471-244X-7-25-4-20", "1471-244X-7-25-4-21", "1471-244X-7-25-4-22", "1471-244X-7-25-4-23", "1471-244X-7-25-4-24", "1471-244X-7-25-4-25", "1471-244X-7-25-4-26", "1471-244X-7-25-4-27", "1471-244X-7-25-4-28", "1471-244X-7-25-4-29", were removed
- 14.04.2016:The test set has been updated. The following changes were made:
- Figure separation: image "1471-2261-9-18-2" was removed
- Multi-label classification: image "1471-2261-9-18-2" was removed
- Subfigure classification: images "1471-2261-9-18-2-1","1471-2261-9-18-2-2", "1471-2261-9-18-2-3", "1471-2261-9-18-2-4", "1471-2148-9-170-4-2 2" and "1471-213X-1-12-1-6 2" were removed
- 02.02.2016: We regret to inform you that we are unable to run the X-rays classification subtask in 2016. However, the medical compound figure separation and multi-label classification tasks will still run
Medical compound figure separation and multi-label classification
Since 2004, ImageCLEF has run the medical task, ImageCLEFmed, to promote research on clinical image systems. ImageCLEFmed has evolved strongly to adapt to current challenges in the domain. The objective of ImageCLEFed is to work on compound figures of the biomedical literature and to separate them if possible and/or attach to the sub parts labels about the content. In addition, a new subtask to predict image captions will be introduced in 2016.
Schedule
- 15.11.2015: registration opens for all ImageCLEF tasks (until 22.04.2016)
- 01.02.2016: development data release starts
- 15.03.2016: test data release starts
- 04.05.2016: deadline for submission of runs by the participants
- 15.05.2016: release of processed results by the task organizers
2528.05.2016: deadline for submission of working notes papers by the participants- 17.06.2016: notification of acceptance of the working notes papers
- 01.07.2016: camera ready working notes papers
- 05.-08.09.2016: CLEF 2016, Évora, Portugal
Citations
- When referring to ImageCLEFmed 2016 task general goals, general results, etc. please cite the following publication:
- Alba García Seco de Herrera, Roger Schaer, Stefano Bromuri and Henning Müller, Overview of the ImageCLEF 2016 medical task, in: CLEF working notes 2016, Évora, Portugal, 2016.
-
BibText:
@InProceedings{GSB2016,
-
Title = {Overview of the {ImageCLEF} 2016 medical task},
Author = {Garc\'ia Seco de Herrera, Alba and Schaer, Roger and Bromuri, Stefano and M\"uller, Henning },
Booktitle = {Working Notes of {CLEF} 2016 (Cross Language Evaluation Forum)},
Year = {2016},
Month = {September},
Location = {\'Evora, Portugal}}
- Jayashree Kalpathy-Cramer, Alba García Seco de Herrera, Dina Demner-Fushman, Sameer Antani, Steven Bedrick and Henning Müller, Evaluating Performance of Biomedical Image Retrieval Systems –an Overview of the Medical Image Retrieval task at ImageCLEF 2004-2014 (2014), in: Computerized Medical Imaging and Graphics
-
BibText:
@Article{KGD2014,-
Title = {Evaluating Performance of Biomedical Image Retrieval Systems-- an Overview of the Medical Image Retrieval task at {ImageCLEF} 2004--2014},
Author = {Kalpathy--Cramer, Jayashree and Garc\'ia Seco de Herrera, Alba and Demner--Fushman, Dina and Antani, Sameer and Bedrick, Steven and M\"uller, Henning},
Journal = {Computerized Medical Imaging and Graphics},
Year = {2014}}
Motivation
An estimated 40% of the figures in PubMed Central are compound figures (images consisting of several sub figures) like the images above. When data of articles is made available digitally, often the compound images are not separated but made available in a single block. Information retrieval systems for images should be capable of distinguishing the parts of compound figures that are relevant to a given query. A major step for making the content of the compound figures accessible is the detection of compound figures and then their separation into sub figures that can subsequently be classified into modalities and made available for research.
As in 2015, the medical compound figure subtask of ImageCLEF 2016 uses a subset of PubMed Central.
Subtask overview
There are five types of subtasks in 2016:
- Compound figure detection:
This subtask was first introduced in 2015. Compound figure identification is a required first step to make available compound images from the literature. Therefore, the goal of this subtask is to identify whether a figure is a compound figure or not. The subtask makes training data available containing compound and non compound figures from the biomedical literature. - Multi-label classification:
This subtask was first introduced in 2015. Characterization of compound figures is difficult, as they may contain subfigures from various imaging modalities or image types. This subtask aims to label each compound figure with each of the modalities (of the 30 classes of a defined hierarchy shown below) of the subfigures contained without knowing where the separation lines are. - Figure separation:
This subtask was first introduced in 2013. The subtask makes available training data with separation labels of the figures and then a test data set where the labels will be made available after the submission of the results. In 2016, a larger number of compound figures is distributed compared to the previous years. - Subfigure classification:
This subtask was first introduced in 2015 but similar to the modality classification subtask organized in 2011-2013. This subtask aims to classify images into the 30 classes of the hierarchy shown below. The images are the subfigures extracted from the compound figures distributed for the figure separation subtask. - Caption prediction:
This is a new subtask that will be introduced in 2016. This subtask aims to define automatic captioning algorithms for the diagnostic images provided as training and testing set. Images and associated captions will be provided.
Classification hierarchy
The following hierarchy is used for the modality classification, same classes as in ImageCLEF 2015.
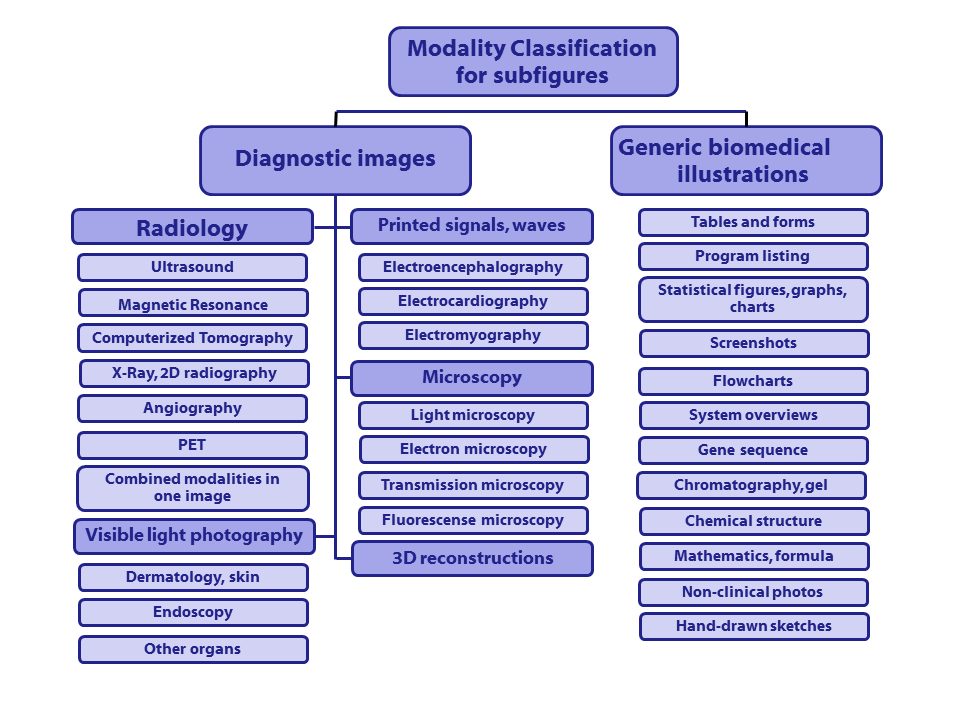
Class codes with descriptions (class codes need to be specified in run files):
([Class code] Description)
- [Dxxx] Diagnostic images:
- [DRxx] Radiology (7 categories):
- [DRUS] Ultrasound
- [DRMR] Magnetic Resonance
- [DRCT] Computerized Tomography
- [DRXR] X-Ray, 2D Radiography
- [DRAN] Angiography
- [DRPE] PET
- [DRCO] Combined modalities in one image
- [DVxx] Visible light photography (3 categories):
- [DVDM] Dermatology, skin
- [DVEN] Endoscopy
- [DVOR] Other organs
- [DSxx] Printed signals, waves (3 categories):
- [DSEE] Electroencephalography
- [DSEC] Electrocardiography
- [DSEM] Electromyography
- [DMxx] Microscopy (4 categories):
- [DMLI] Light microscopy
- [DMEL] Electron microscopy
- [DMTR] Transmission microscopy
- [DMFL] Fluorescence microscopy
- [D3DR] 3D reconstructions (1 category)
- [Gxxx] Generic biomedical illustrations (12 categories):
- [GTAB] Tables and forms
- [GPLI] Program listing
- [GFIG] Statistical figures, graphs, charts
- [GSCR] Screenshots
- [GFLO] Flowcharts
- [GSYS] System overviews
- [GGEN] Gene sequence
- [GGEL] Chromatography, Gel
- [GCHE] Chemical structure
- [GMAT] Mathematics, formulae
- [GNCP] Non-clinical photos
- [GHDR] Hand-drawn sketches
Data collection
The dataset used in this task is a subset of images contained in articles from the biomedical literature extracted from the PubMed Central.
- Compound figure detection:
21,000 images labelled as compound figures or not compound figures will be distributed. Images captions will be also provided. - Figure separation:
A subset of the compound figures from the compound figure detection task will be distributed to be separated into subfigures. Images captions will be also provided. - Multi-label classification:
A subset of the compound figures are distributed for the multi-label task. Images captions will be also provided. - Subfigure classification:
Figures from the multi-label classification task are separated into subfigures and each of the subfigures are labelled.
If the figure ID is "1297-9686-42-10-3", then the corresponding subfigures IDs are "1297-9686-42-10-3-1", "1297-9686-42-10-3-2", "1297-9686-42-10-3-3" and "1297-9686-42-10-3-4" - Caption prediction:
The data disclosed to the participants will involve 10,000 images from diagnostic category and relative captions.
The testing set will comprise other 10,000 diagnostic images, but the captions will not be included. The name of the
folders contaning the data are FigurePredictionTrain2016 for the training dataset and FigurePredictionTest2016 for the testing
dataset.
Submission instructions
Please note that each group is allowed a maximum of 10 runs per subtask.
Each runfile's name has to start with the abbreviation of the subtask. Here more specifically:
Example of valid runfile names: cp.run1.txt, FsRun2.txt, FDxxxx, MLC-method1.run,...
Compound figure detection
The format of the result submission for the Compound figure detection subtask should be the following:
1471-2458-10-S1-S4-3 COMP 0.9
1471-2458-10-52-5 COMP 1
1471-2458-11-133-1 COMP 0.4
1423-0127-17-34-8 NOCOMP 0.8
1465-9921-6-21-6 NOCOMP 0.9
...
where:
- The first column contains the figure-ID (IRI). This ID does not contain the file format ending and it should not represent a file path.
- The second column is the classcode.
- The third column represents the normalized score (between 0 and 1) that your system assigned to the specific result.
You need to respect the following constraints:
- Each specified figure has to be part of the collection (dataset).
- A figure cannot be contained more than once.
- All figures of the test set have to be contained in the runfile.
- Only known class codes are accepted.
Please note that each group is allowed to submit a maximum of 10 runs.
Compound figure separation
The format of the result submission for the compound figure separation subtask has to be an XML file with the following structure :
where:
- The root element is <annotations>.
- The root contains one <annotation> element per image. Each one of these elements has to contain :
- A <filename> element with the name of the compound image (IRI) (excluding the file extension)
- One or more <object> elements that define the bounding box of each subfigure in the image. Each <object> must contain :
- 4 <point> elements that define the 4 corners of the bounding box. The <point> elements must have two attributes (x and y), which correspond to the horizontal and vertical pixel position, respectively. The preferred order of the points is :
- top-left
- top-right
- bottom-left
- bottom-right
You also need to respect the following constraints:
- Each specified image has to be part of the collection (dataset).
- An Image cannot appear more than once in a single XML results file.
- All the images of the testset must be contained in the runfile.
- The resulting XML file MUST validate against the XSD schema that will be provided.
Multi-label classification
The format of the result submission for the Multi-label classification classification subtask should be the following:
1751-0147-52-24-3 DRMR DRXR
1475-925X-6-10-8 DRUS GHDR
1471-2210-10-7-3 GCHE
1475-2875-6-10-2 GFIG
...
where:
- The first column contains the figure-ID (IRI). This ID does not contain the file format ending and it should not represent a file path.
- The rest of the columns contain the classcode.
You need to respect the following constraints:
- Each specified figure has to be part of the collection (dataset).
- A figure cannot be contained more than once.
- All figures of the test set have to be contained in runfile.
- Only known classcodes are accepted.
Please note that each group is allowed a maximum of 10 runs.
Subfigure classification
Similar to the compound figure detection subtask, the format of the result submission for the Subfigure classification subtask should be the following:
1743-422X-4-12-1-4 D3DR 0.9
1471-2156-8-36-3-4 GFIG 1
1475-2859-9-86-6-1 GFIG 0.4
1475-2840-10-59-2-4 DMLI 0.8
...
where:
- The first column contains the subfigure-ID (IRI). This ID does not contain the file format ending and it should not represent a file path.
- The second column is the classcode.
- The third column represents the normalized score (between 0 and 1) that your system assigned to the specific result.
You need to respect the following constraints:
- Each specified subfigure has to be part of the collection (dataset).
- A subfigure cannot be contained more than once.
- All subfigures of the test set have to be contained in runfile.
- Only known class codes are accepted.
Caption prediction
For the submission of the caption prediction task we expect the following format:
- 1743-422X-4-12-1-4 description of the first image in one single line
- 1743-422X-4-12-1-3 description of the second image....
- 1743-422X-4-12-1-2 descrition of the third image...
You need to respect the following constraints:
- Each specified test figure has to be part of the collection (dataset).
- You should not include special characters in the description.
Evaluation methodology
- Compound figure detection: will be evaluated based on classification accuracy.
- Multi-label classification: will be evaluated based the Hamming Loss and f-measure.
- Compound Figure Separation : The Java Archive (JAR) containing the application to run the evaluation can be downloaded below. The source code of the application is also contained in the ZIP archive.
Download Compound Figure Separation Evaluation Tool & Source
- Subfigure classification: will be evaluated based on classification accuracy.
- Caption prediction: We will use a set of evaluation metrics taking into consideration the similarity between the predicted text and the real text:
update: to limit the complexity of the task, we will use the cosine similarity between the predicted caption and the real caption, after removing
stop words. - Alba García Seco de Herrera<albagarcia@nih.gov>, National Library of Medicine (NLM/NIH), Bethesda, MD, USA
- Stefano Bromuri <stefano.bromuri@ou.nl>Open Universiteit, Netherlands, Heerlen
- Roger Schaer <roger.schaer@hevs.ch>, University of Applied Sciences Western Switzerland, Sierre, Switzerland
- Henning Müller <henning.mueller@hevs.ch>, University of Applied Sciences Western Switzerland, Sierre, Switzerland
| Compound figure detection |
| Group name | Run | Run type | Correctly classified in % |
| DUTIR | CFD_DUTIR_Mixed_AVG.txt | mixed | 92.70 |
| CIS UDEL | CFDRun10.txt | mixed | 90.74 |
| CIS UDEL | CFDRun09.txt | mixed | 90.39 |
| CIS UDEL | CFDRun05.txt | mixed | 90.39 |
| CIS UDEL | CFDRun07.txt | mixed | 85.47 |
| CIS UDEL | CFDRun8.txt | mixed | 69.06 |
| CIS UDEL | CFDRun06.txt | mixed | 52.25 |
| CIS UDEL | CFDRun01.txt | textual | 85.47 |
| DUTIR | CFD_DUTIR_Textual_RNN.txt | textual | 86.05 |
| DUTIR | CFD_DUTIR_Textual_CNN.txt | textual | 87.03 |
| MLKD | CFD2.txt | textual | 88.13 |
| DUTIR | CFD_DUTIR_Visual_CNNs.txt | visual | 92.01 |
| CIS UDEL | CFDRun04.txt | visual | 89.64 |
| CIS UDEL | CFDRun02.txt | visual | 89.29 |
| CIS UDEL | CFDRun03.txt | visual | 69.82 |
| Multi-label classification |
| Group name | Run | Hamming Loss | f-measure |
| BMET | MLC-BMET-multiclass-test-max-all | 0.0131 |
0.295 |
| BMET | MLC-BMET-multiclass-test-prob-max-all | 0.0135 |
0.32 |
| MLKD | MLC2 | 0.0294 | 0.32 |
| Figure separation |
| Group name | Run | Run type | Correctly classified in % |
| CIS UDEL | FS.run9.xml | visual | 84.43 |
| CIS UDEL | FS.run7.xml | visual | 84.08 |
| CIS UDEL | FS.run6.xml | visual | 84.03 |
| CIS UDEL | FS.run8.xml | visual | 83.04 |
| CIS UDEL | FS.run5.xml | visual | 81.23 |
| CIS UDEL | FS.run3.xml | visual | 75.27 |
| CIS UDEL | FS.run4.xml | visual | 74.83 |
| CIS UDEL | FS.run2.xml | visual | 74.30 |
| CIS UDEL | FS.run1.xml | visual | 73.57 |
| Subfigure classification |
| Group name | Run | Run type | Correctly classified in % |
| BCSG | SC_BCSG_run10_Ensemble_Vote.txt | mixed | 88.43 |
| BCSG | SC_BCSG_run5_Mixed_DeCAF-ResNet-152.txt | mixed | 88.21 |
| BCSG | SC_BCSG_run4_Hierarchy.txt | mixed | 87.61 |
| BCSG | SC_BCSG_run3_Mixed.txt | mixed | 87.56 |
| BCSG | SC_BCSG_run9_Mixed_NMF.txt | mixed | 86.96 |
| BCSG | SC_BCSG_run6_LateFusion.txt | mixed | 84.44 |
| BCSG | SC_BCSG_run2_Textual.txt | textual | 72.22 |
| MLKD | SC2.txt | textual | 58.37 |
| BCSG | SC_BCSG_run8_DeCAF_ResNet-152_PseudoInverse.txt | visual | 85.38 |
| BCSG | SC_BCSG_run1_Visual.txt | visual | 84.46 |
| IPL | SC_enriched_GBOC_1x1_256_RGB_Phow_Default_1500_EarlyFusion.txt | visual | 84.01 |
| IPL | SC_enriched_GBOC_1x1_128_HSV_Phow_RGB_1500_EarlyFusion.txt | visual | 83.46 |
| IPL | SC_enriched_GBOC_1x1_128_HSV_Phow_RGB_1500_LateFusion.txt | visual | 82.66 |
| IPL | SC_enriched_GBOC_1x1_128_RGB_Phow_Default_1500_LateFusion.txt | visual | 82.50 |
| IPL | SC_original_GBOC_1x1_256_RGB_w_0.6_Phow_Default_1500_w_0.4_EarlyFusion.txt | visual | 81.73 |
| IPL | SC_original_GBOC_1x1_256_RGB_Phow_Default_1500_EarlyFusion.txt | visual | 8170 |
| IPL | SC_original_GBOC_1x1_128_RGB_Phow_Default_1500_EarlyFusion.txt | visual | 81.32 |
| BCSG | SC_BCSG_run7_GoogLeNet-PReLU-Xavier.txt | visual | 81.03 |
| IPL | SC_original_GBOC_1x1_256_RGB_Phow_Default_1500_LateFusion.txt | visual | 80.17 |
| IPL | SC_original_GBOC_1x1_128_HSV_Phow_RGB_1500_LateFusion.txt | visual | 80.14 |
| IPL | SC_original_GBOC_1x1_128_RGB_Phow_Default_1500_LateFusion.txt | visual | 79.45 |
| BMET | SC-BMET-subfig-test-prob-sum.txt | visual | 77.55 |
| BMET | SC-BMET-subfig-test-score-sum-merged.txt | visual | 77.53 |
| BMET | SC-BMET-subfig-test-score-sum-cropscale.txt | visual | 77.50 |
| BMET | SC-BMET-subfig-test-majority.txt | visual | 77.26 |
| BMET | SC-BMET-subfig-test-prob-max.txt | visual | 76.38 |
| NWPU | sc.run3.txt | visual | 74.17 |
| NWPU | sc.run4.txt | visual | 74.14 |
| NWPU | sc.run5.txt | visual | 73.97 |
| CIS UDEL | SCRun1.txt | visual | 72.46 |
| CIS UDEL | SCRun2.txt | visual | 71.53 |
| NWPU | sc.run2.txt | visual | 71.41 |
| NWPU | sc.run1.txt | visual | 71.19 |
| CIS UDEL | SCRun4.txt | visual | 68.69 |
| CIS UDEL | SCRun3.txt | visual | 68.17 |
| NOVASearch | SC_NOVASearch_cnn_10_dropout_vgglike.run | visual | 65.31 |
| NOVASearch | SC_NOVASearch_cnn_8_vgglike.run | visual | 65.17 |
| NOVASearch | SC_NOVASearch_cnn_prelu.run | visual | 63.80 |
| NOVASearch | SC_NOVASearch_cnn_10_vgglike.run | visual | 63.29 |
| CIS UDEL | SCRun7.txt | visual | 53.24 |
| CIS UDEL | SCRun6.txt | visual | 53.16 |
| CIS UDEL | SCRun5.txt | visual | 15.62 |






