- ImageCLEF 2025
- LifeCLEF 2025
- ImageCLEF 2024
- LifeCLEF 2024
- ImageCLEF 2023
- LifeCLEF 2023
- ImageCLEF 2022
- LifeCLEF2022
- ImageCLEF 2021
- LifeCLEF 2021
- ImageCLEF 2020
- LifeCLEF 2020
- ImageCLEF 2019
- LifeCLEF 2019
- ImageCLEF 2018
- LifeCLEF 2018
- ImageCLEF 2017
- LifeCLEF2017
- ImageCLEF 2016
- LifeCLEF 2016
- ImageCLEF 2015
- LifeCLEF 2015
- ImageCLEF 2014
- LifeCLEF 2014
- ImageCLEF 2013
- ImageCLEF 2012
- ImageCLEF 2011
- ImageCLEF 2010
- ImageCLEF 2009
- ImageCLEF 2008
- ImageCLEF 2007
- ImageCLEF 2006
- ImageCLEF 2005
- ImageCLEF 2004
- ImageCLEF 2003
- Publications
- Old resources
You are here
FungiCLEF 2024

Schedule
- December 2023: Registration opens for all LifeCLEF challenges Registration is free of charge
- 13 March 2024: Competition Start
- 24 May 2024: Competition Deadline
- 31 May 2024: Deadline for submission of working note papers by participants [CEUR-WS proceedings]
- 21 June 2024: Notification of acceptance of working note papers [CEUR-WS proceedings]
- 8 July 2024: Camera-ready deadline for working note papers.
- 9-12 Sept 2024: CLEF 2024 Grenoble - France
All deadlines are at 11:59 PM CET on a corresponding day unless otherwise noted. The competition organizers reserve the right to update the contest timeline if they deem it necessary.
Results
All teams that provided a runnable code were scored, and their scores are available on HuggingFace.
Official competition results are listed in the Private and Public leaderboards, respectively.
- The 1st place team: IES (Stefan Wolf, Philipp Thelen and Jürgen Beyerer)
- The 2nd place team: jack-etheredge(Jack Etheredge)
- The 3rd place team: upupup (Bao-Feng Tan, Yang-Yang Li, Peng Wang, Lin Zhao and Xiu-Shen Wei)
Private Leaderboard Evaluation
(orange = baseline)
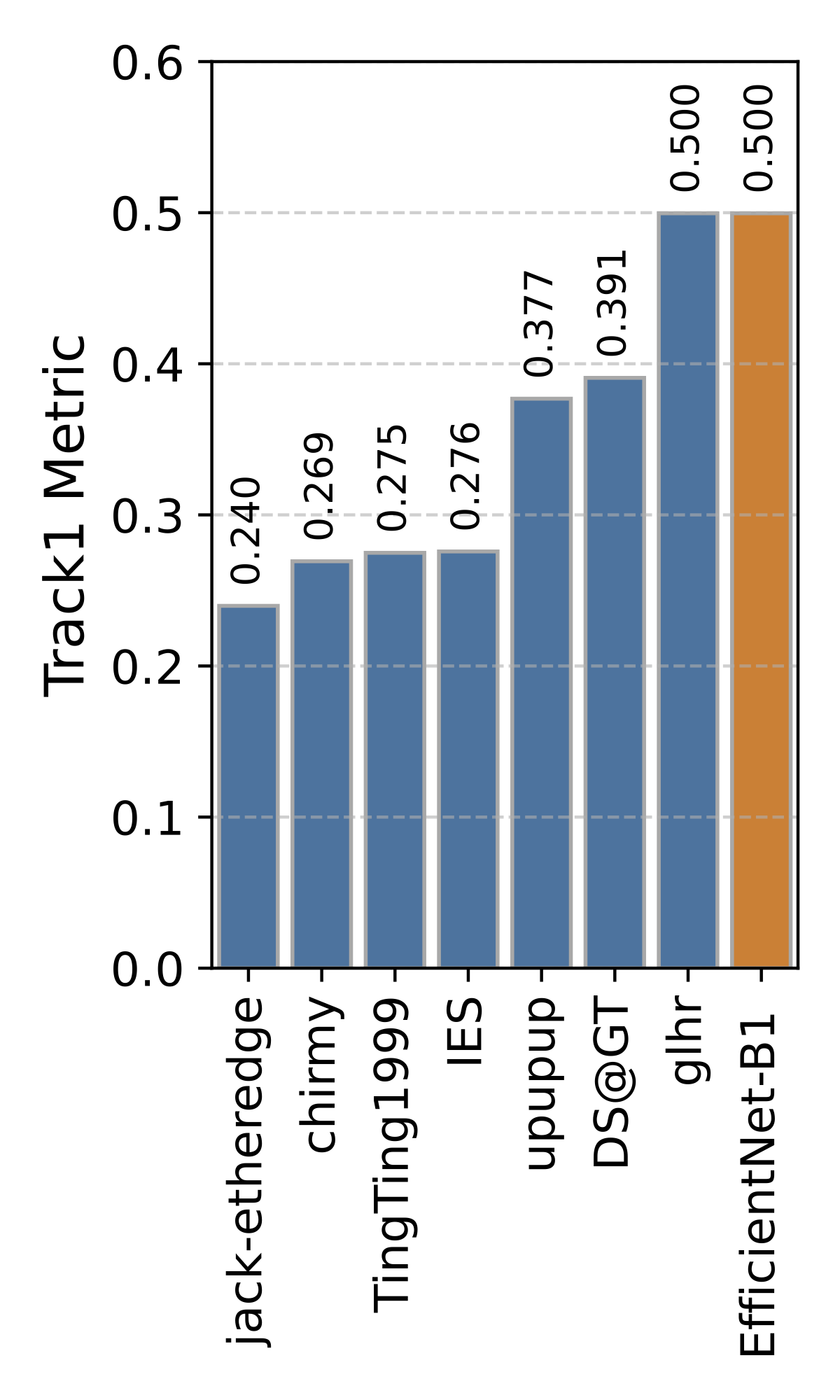
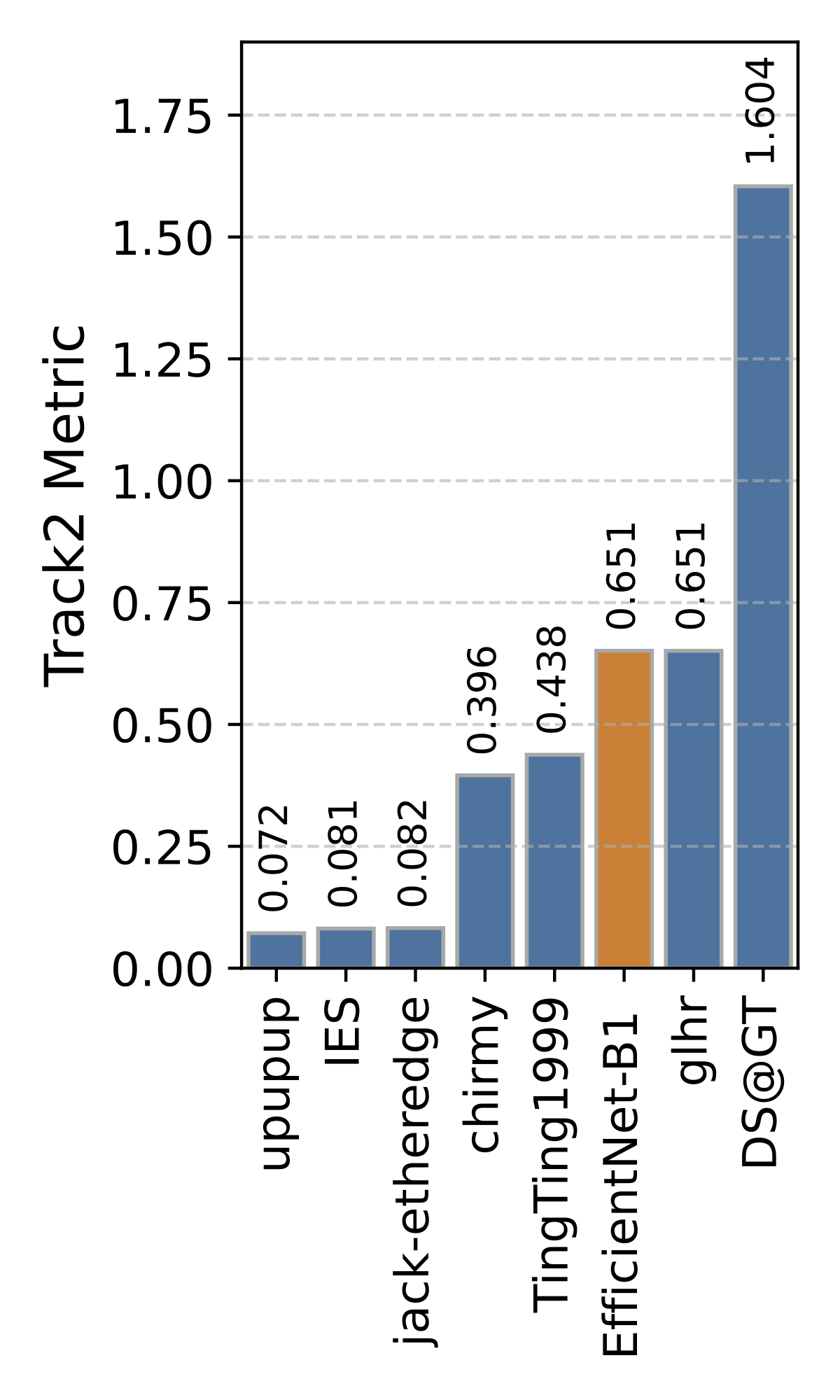
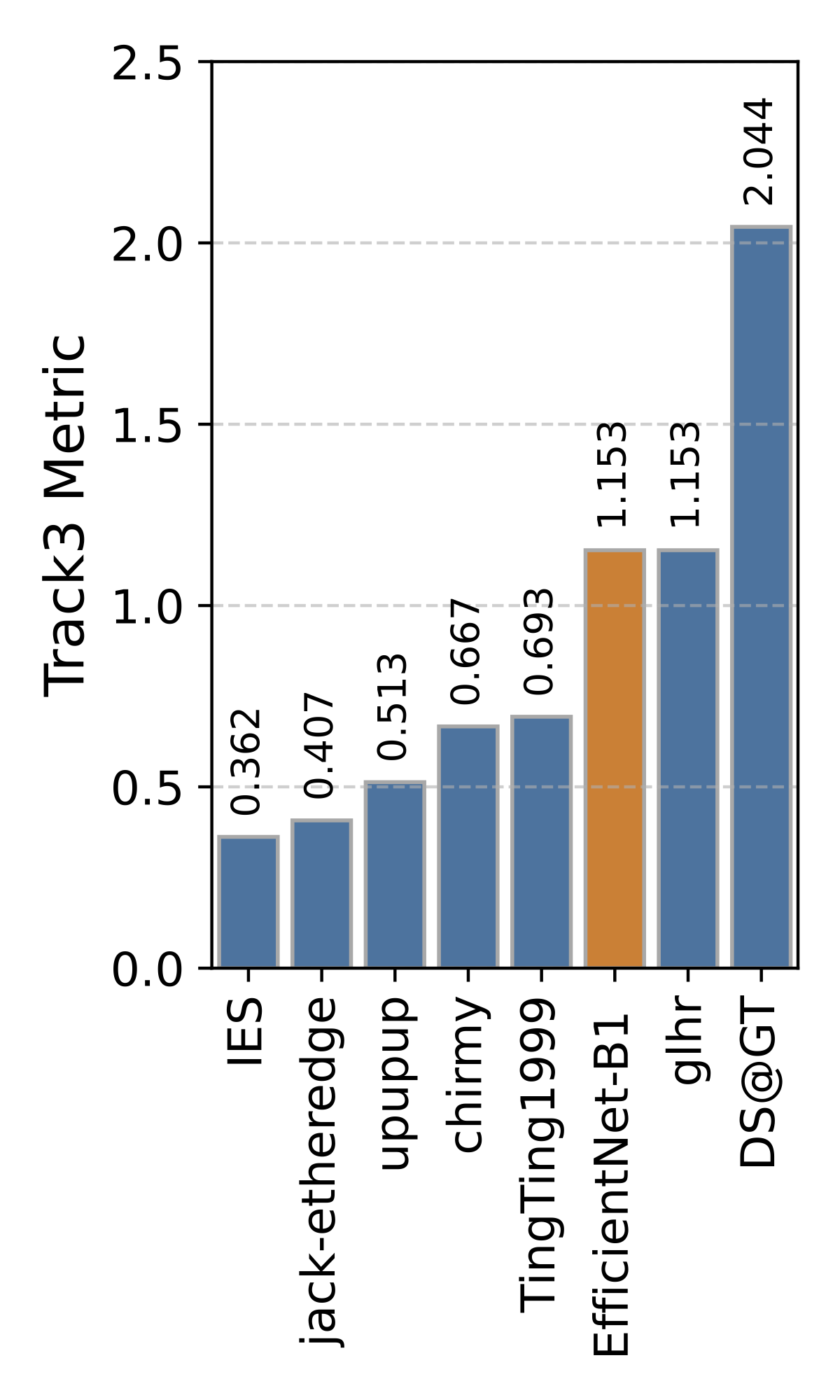
Motivation
Automatic recognition of fungi species aids mycologists, citizen scientists, and nature enthusiasts in identifying species in the wild while supporting the collection of valuable biodiversity data. To be effective on a large scale, such as in popular citizen science projects, it needs to efficiently predict species with limited resources and handle many classes, some of which have just a few recorded observations. Additionally, rare species are often excluded from training, making it difficult for AI-powered tools to recognize them. Based on our measurements, we recognized that about 20\% of all verified observations (20,000) involve rare or under-recorded species, highlighting the need to accurately identify these species.
Task Description
Given the set of real fungi species observations and corresponding metadata, the goal of the task is to create a classification model that returns a ranked list of predicted species for each observation (multiple photographs of the same individual + geographical location).
The classification model must fit limits for memory footprint and a prediction time limit (120 minutes) within a given HuggingFace server instance (Nvidia T4 small 4vCPU, 15GB RAM, 16GB VRAM).
Note: Since the test set contains multiple out-of-the-scope classes. The solution has to handle such classes.
Participation requirements
- 1. Subscribe to CLEF (LifeCLEF – FungiCLEF2024 task) by filling out this form.
- 2. Submit your solution before the competition deadline. More info on a HuggingFace competition platform.
Publication Track
All registered participants are encouraged to submit a working-note paper to peer-reviewed LifeCLEF proceedings (CEUR-WS) after the competition ends.
This paper must provide sufficient information to reproduce the final submitted runs.
Only participants who submitted a working-note paper will be part of the officially published ranking used for scientific communication.
The results of the campaign appear in the working notes proceedings published by CEUR Workshop Proceedings (CEUR-WS.org).
Selected contributions among the participants will be invited for publication in the Springer Lecture Notes in Computer Science (LNCS) the following year.
For detailed instructions, please refer to SUBMISSION INSTRUCTIONS.
A summary of the most important points:
- All participating teams with at least one graded submission, regardless of the score, should submit a CEUR working notes paper.
- Submission of reports is done through EasyChair – please make absolutely sure that the author (names and order), title, and affiliation information you provide in EasyChair match the submitted PDF exactly
- Deadline for the submission of initial CEUR-WS Working Notes Papers (for the peer-review process): 31 May 2024
- Deadline for the submission of Camera Ready CEUR-WS Working Notes Papers:: 8 July 2024
- Templates are available here
- Working Notes Papers should cite both the LifeCLEF 2024 overview paper as well as the PlantCLEF task overview paper, citation information will be added in the Citations section below as soon as the titles have been finalized.
Context
This competition is held jointly as part of:
- the LifeCLEF 2024 lab of the CLEF 2024 conference, and of
- the FGVC11 workshop, organized in conjunction with CVPR 2024
Data
The challenge dataset is primarily based on the Danish Fungi 2020 dataset, which contains 295,938 training images belonging to 1,604 species observed mostly in Denmark. All training samples passed an expert validation process, guaranteeing high-quality labels. Rich observation metadata about habitat, substrate, time, location, EXIF, etc., are provided.
The validation set contains 30,131 observations with 60,832 images and 2,713 species, covering the whole year and including observations collected across all substrate and habitat types.
Using additional data or metadata is permitted!
| Image Data | Metadata |
Evaluation process
As last year, we will calculate four custom metrics and macro averaged F1 Score and Accuracy. All "unusual metrics" are explained on the competition website. The code is provided on GitHub.
The FungiCLEF 2024 recognition challenge is running with several metrics representing different decision problems, where the goal is to minimize the average empirical loss \( L \) for decisions \(q(x)\) over observations \(x\) and true labels \(y\), given a cost function \(W (y, q(x)) \).
\[ L = \dfrac{1}{n} \sum_{i=1}^n W (k_i, q(x_i)) \]
1. Standard Classification with "unknown" category. The first metric is the standard classification error, i.e. the average error of the predicted class.
All species not represented in the training set should correctly be classified as an "unknown" category.
The decision function is simple: for each observation is simply represented by an identity matrix, i.e.
\[ W_1(y,q(x))) = \left\{
\begin{matrix}
0 & \text{ if } q(x) = y \\
1 & \text{ otherwise}
\end{matrix}
\right.
\]
2. Cost for confusing edible species for poisonous and vice versa.
Let us have a function \(d\) that indicates dangerous (poisonoius) species as \(d(y)=1\) if species \( y \) is poisonois, and \(d(y)=0\) otherwise.
Let us denote \(c_\text{PSC}\) the cost for poisonous species confusion (if a poisonous observation was misclassified as edible) and \(c_\text{ESC}\) the cost for edible species confusion (if an edible observation was misclassified as poisonous).
\[ W_2(y,q(x))) = \left\{
\begin{matrix}
0 & \text{if } d(y) = d(q(y)) \hfill \\
c_\text{PSC} & \text{if } d(y) = 1 \text{ and } d(q(y)) = 0 \\
c_\text{ESC} & \text{otherwise} \hfill
\end{matrix}
\right.
\]
For the benchmark, we set \(c_\text{ESC} = 1\) and \(c_\text{PSC}=100\).
3. A user-focused loss composes of both the classification error and the poisonous/edible confusion.
\[ L_3 = \sum_i W_1 (k_i, q(x_i)) + W_2 (k_i, q(x_i)) \]
Other resources
For more FungiCLEF-related info, please refer to overview papers from previous editions. Besides, you can check out other competitions from the CLEF-LifeCLEF and CVPR-FGVC workshops.
- Papers describing the Danish Fungi dataset and our Fungi Recognition App.
- FungiCLEF2023 Overview Paper, and FungiCLEF2022 Overview Paper
- CVPR - FGVC workshop page for more information about the FGVC11 workshop.
- LifeCLEF page for more information about LifeCLEF challenges and working notes submission procedure.
Organizers
- Lukas Picek, INRIA--Montpellier, France & Dept. of Cybernetics, FAV, University of West Bohemia, Czechia, lukaspicek@gmail.com
- Milan Sulc, Czechia, milansulc01@gmail.com
- Jiri Matas, The Center for Machine Perception Dept. of Cybernetics, FEE, Czech Technical University, Czechia, matas@cmp.felk.cvut.cz
- Jacob Heilmann-Clausen, Center for Macroecology, Evolution and Climate; University of Copenhagen, Denmark, jheilmann-clausen@sund.ku.dk
Machine Learning
Mycology
Credits





Acknowledgement
This workshop was supported by the Technology Agency of the Czech Republic, project No. SS05010008 and project No. SS73020004.


