- ImageCLEF 2025
- LifeCLEF 2025
- ImageCLEF 2024
- LifeCLEF 2024
- ImageCLEF 2023
- LifeCLEF 2023
- ImageCLEF 2022
- LifeCLEF2022
- ImageCLEF 2021
- LifeCLEF 2021
- ImageCLEF 2020
- LifeCLEF 2020
- ImageCLEF 2019
- LifeCLEF 2019
- ImageCLEF 2018
- LifeCLEF 2018
- ImageCLEF 2017
- LifeCLEF2017
- ImageCLEF 2016
- LifeCLEF 2016
- ImageCLEF 2015
- LifeCLEF 2015
- ImageCLEF 2014
- LifeCLEF 2014
- ImageCLEF 2013
- ImageCLEF 2012
- ImageCLEF 2011
- ImageCLEF 2010
- ImageCLEF 2009
- ImageCLEF 2008
- ImageCLEF 2007
- ImageCLEF 2006
- ImageCLEF 2005
- ImageCLEF 2004
- ImageCLEF 2003
- Publications
- Old resources
You are here
Plant Identification 2013

| News |
| 17.10.2013 A direct link to the working note about the overview of the task: The ImageCLEF 2013 plant image identification task, CLEF 2013 working notes, Goëau H., Bonnet P., Joly A., Bakic V., Barthélémy D., Boujemaa N., Molino J-F., ImageCLEF 2013 working notes, Valencia, Spain. |
| 14.10.2013 The public packages containing all the data of the ImageCLEF 2013 plant retrieval task are now available (including the ground truth, an executable in order to compute new scores, the working notes and the oral and poster presentations at ImageCLEF 2012 Workshop, and some additional informations). The entire data is under Creative Common license. You can download it using the following urls: https://lab.plantnet.org/LifeCLEF/PlantCLEF2013/ImageCLEF2013PlantTaskTe... https://lab.plantnet.org/LifeCLEF/PlantCLEF2013/ImageCLEF2013PlantTaskTr... https://lab.plantnet.org/LifeCLEF/PlantCLEF2013/ImageCLEF2013PlantTaskTr... |
Context
If agricultural development is to be successful and biodiversity is to be conserved, then accurate knowledge of the identity, geographic distribution and uses of plants is essential. Unfortunately, such basic information is often only partially available for professional stakeholders, teachers, scientists and citizens, and often incomplete for ecosystems that possess the highest plant diversity. So that simply identifying plant species is usually a very difficult task, even for professionals (such as farmers or wood exploiters) or for the botanists themselves. Using image retrieval technologies is nowadays considered by botanists as a promising direction in reducing this taxonomic gap. Evaluating recent advances of the IR community on this challenging task might therefore have a strong impact. The organization of this task is funded by the French project Pl@ntNet (INRIA, CIRAD, Telabotanica) and supported by the European Coordination Action CHORUS+.
Task Overview
Following the success of ImageCLEF 2011 and ImageCLEF 2012 plant identification tasks, we are glad to organize this year a new challenge dedicated to botanical data. This year, the task will be focused on tree and herb species identification, based on different types of images. Main novelties compared to last year are the following:
- more species: the number of species will be this year about 250, which is an important step towards covering the entire flora of a given region.
- multi-view plant retrieval vs. leaf-based retrieval: query and test pictures will now cover different organs (or views) of the individual plants and not only their leaves.
The leaf is actually far from being the only useful organ for accurate identification. As an example, the 6 species depicted in the figure below share the same French common name of laurel ("laurier") in spite of even though they belong to different taxonomic groups (5 genera, 5 families). In this case, while it is difficult to identify the species with more or less the same-sized elliptic shaped leaves, it is indisputably easier with the flowers.

6 different species sharing the same common name
The training and test data will be composed of images collected through a citizen sciences initiative that was initiated 2 years ago in collaboration with Telabotanica (social network of amateur and expert botanists). This makes the task closer to the conditions of a real-world application: (i) images of the same species are coming from distinct trees living in distinct areas (ii) pictures are taken by different users that might not used the same protocol to acquire the images (iii) pictures are taken at different periods in the year.
Additional information will include contextual meta-data (author, date, locality name) and some EXIF data.
Dataset
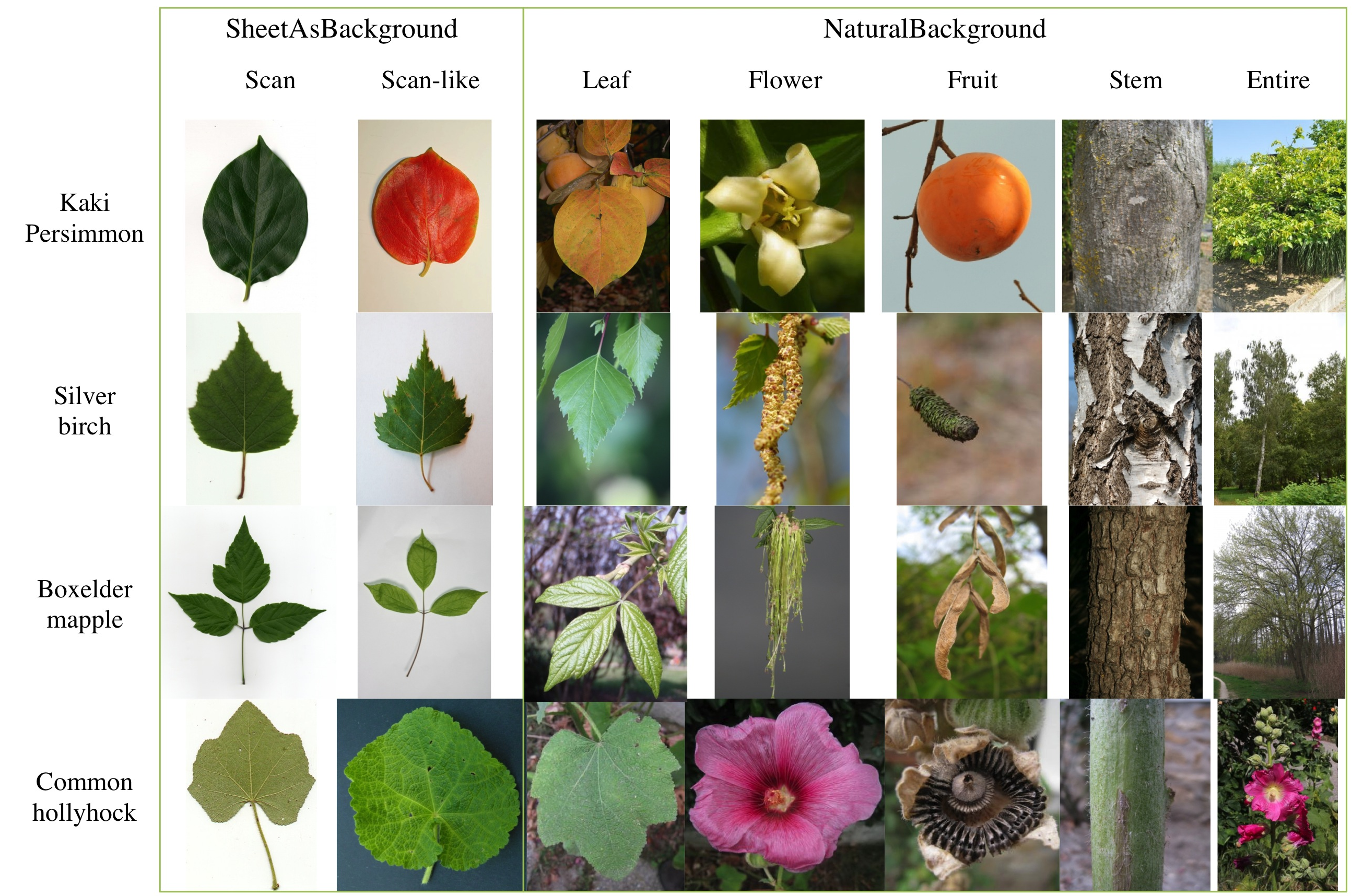
The task will be based on the Pl@ntView dataset which focuses on 250 herb and tree species from France area. It contains 26077 pictures belonging each to one of the 2 following categories:
- SheetAsBackground (or uniform background) (42%): exclusively pictures of leaves in front of a white or colored uniform background produced with a scanner or a camera with a sheet.
- NaturalBackground (for most of the time cluttered natural background") (58%): free natural photographs of different views on different subparts of a plant into the wild.
These 2 main categories can be subdivided into 2 and 5 sub-categories:
- SheetAsBackground Scan (22%): scan of a single leaf.
- SheetAsBackground Scan-like (22%): photograph of a single leaf in front of an uniform artificial background.
- NaturalBackground Leaf (16%): photograph of one leaf or more directly on the plant or near the plant on the floor or other non uniform background.
- NaturalBackground Flower (18%): photograph of one flower or a group of flowers (inflorescence) directly on the plant.
- NaturalBackground Fruit (8%): photograph of one fruit or a group of fruits (infructescence) directly on the plant.
- NaturalBackground Stem (8%): photograph of the stalk, or a stipe, or the bark of the trunk or a main branch of the plant.
- NaturalBackground Entire (8%): photograph of the entire plant, from the floor to the top.
The following figure provides examples of the 2 categories and 7 sub-categories:
These information of categories are reported into the meta-data, in a xml file (one per image) with explicit tags:
- Acquisition Type: "SheetAsBackground" for scan or scan-like pictures of a leaf, "NaturalBackground" for photograph with a cluttered background
- View Content: Leaf, Flower, Fruit, Stem, Entire
Another important additional information in the meta-data is IndividualPlantId, an unique identifier for one single individual plant observed by one same person the same day with the same device with the same lightening conditions. Each individual plant observed may thus be associated to several pictures. It means that these pictures are strongly connected, and may be very similar, even being near-duplicate pictures. As a consequence, pictures with the same IndividualPlantId can not be split across subsets during the identification task like training or test subsets, because they belong to one same indivisible event.
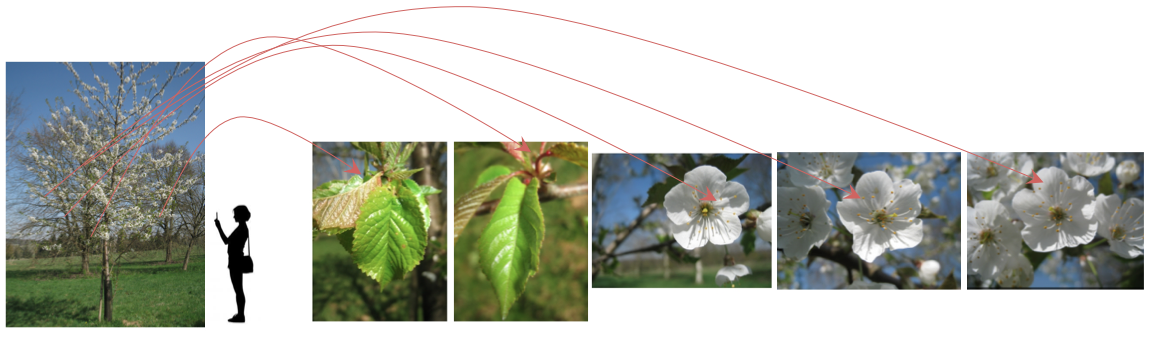
One individual-plant observed the same day by a same author involving several pictures with the same IndividualPlantId.
To sum up each image is associated with the following meta-data:
- Acquisition Type: Uniform ("SheetAsBackground") or Cluttered ("NaturalBackground"),
- View Content: Leaf, Flower, Fruit, Stem, Entire
- IndividualPlantId: the plant id from which several pictures may be associated (see just above-written)
- Taxon: the full taxon names (species, genus, family…) following as possible the most recent and complet information
- ClassId: the class label that must be used as ground-truth. It is a non-official short name of the taxon used for easily designating a taxon (most of the time the species and genus names without the name of the author of the taxon)
- VernacularName: the english common name(s) ,
- Date
- Locality: locality name (a district or a country division or a regions),
- GPSLocality: GPS coordinates of the locality where the plant was observed,
- Author: name of the author of the picture,
- Organization: name of the organization of the author.
And if the image was included in previous plant task:
- Year: ImageCLEF2011 or ImageCLEF2012,
- IndividualPlantId2012: the plant id used in 2012,
- ImageID2012: the image id.jpg used in 2012.
We provide here a set of 2 images and associated xml files (click on a picture):


Partial meta-data information can be found in the image's EXIF, and might include:
- the camera or the scanner model,
- the image resolutions and the dimensions,
- for photos, the optical parameters, the white balance, the light measures…
All data are published under a creative commons license.
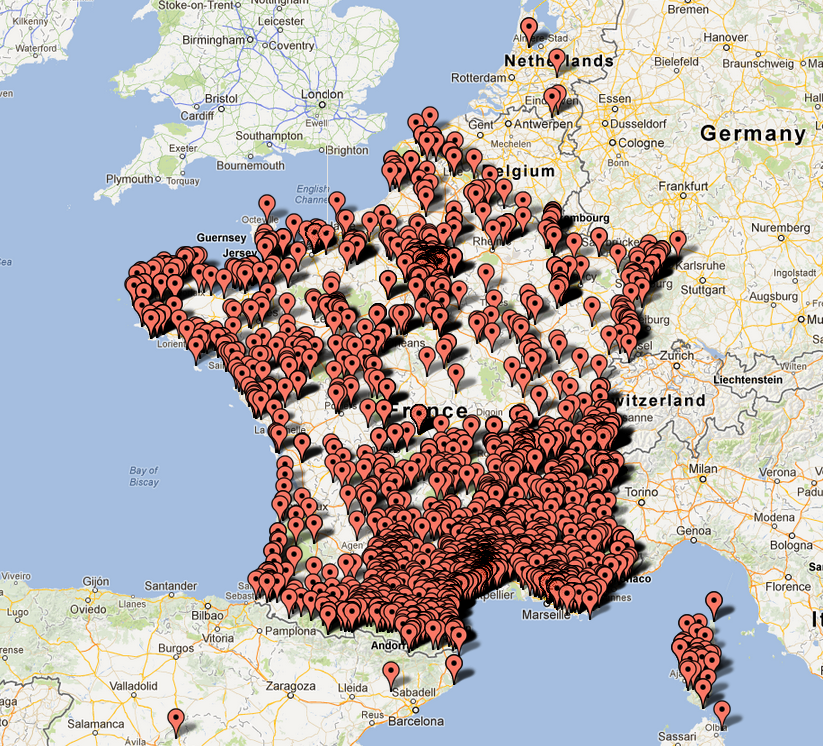
Localities in the ImageCLEF 2013 Plant Task dataset
Task description
The task will be evaluated as a plant species retrieval task. Test images will be divided into two subtasks following the two main category SheetAsBackground and NaturalBackground involving distinct scores like as previous years.
Goal
The goal of the task is to retrieve the correct plant species among the top k species of a ranked list of returned species for each test image. Each participant is allowed to submit up to 4 runs built from different methods. Semi-supervised and interactive approaches, particularly for segmenting leaves from the background, are allowed but will be compared independently from fully automatic methods. Any human assistance in the processing of the test queries has therefore to be signaled in the submitted runs (see next section on how to do that).
training and test data
A part of Pl@ntView dataset will be provided as training data whereas the remaining part will be used later as test data. Training AND test pictures of leaves used during ImageCLEF 2012 art part of the Pl@ntView dataset and they will be systematically included in the training set. New scans and scan-like images of leaves will be introduced and used as test images. rFor the other new views of flower, fruit, stem and entire plant, test pictures will be chosen by randomly sampling 1/3 of the individual plants of each species and removing the pictures with less than 2 training images of the same view type and species in the training data.
- The training data finally results in 20985 images (9781 scans and scan-like pictures of leaf with a "SheetAsBackground", 11204 photographs of "natural background" more precisely 3522 "flower", 2080 "leaf", 1455 "entire", 1387 "fruit", 1337 "stem") with complete xml files associated to them. A ground-truth file listing all images of each species will be provided complementary. Download link of training data will be sent to participants on 29/01/2013.
- The test data results in 5092 images (1250 scans and scan-like pictures of leaf with a "SheetAsBackground", 3842 photographs of "natural background" more precisely 1233 "flower", 790 "leaf", 694 "entire", 520 "fruit", 605 "stem") with purged xml files (i.e without the taxon information that has to be predicted).
run format
The run file must be named as "teamname_runX.run" where X is the identifier of the run (i.e. 1,2 or 3). The run file has to contain as much lines as the total number of predictions, with at least one prediction per test image and a maximum of 250 predictions per test image (250 being the total number of species). Each prediction item (i.e. each line of the file) has to respect the following format :
<test_image_name.jpg;ClassId;rank;score>
The ClassId is the pair <Genus_name_without_author_name Species_name_without_author_name> and forms a unique identifier of the species. These strings have to respect the format provided in the ground-truth file provided with training set (i.e. the same format as the fields <ClassId> in the xml metadata files, see examples in previous section). <rank> is the ranking of a given species for a given test image. <Score> is a confidence score of a prediction item (the lower the score the lower the confidence). Here is a fake run example respecting this format:
myteam_run2.txt
The order of the prediction items (i.e. the lines of the run file) has no influence on the evaluation metric, so that contrary to our example prediction items might be sorted in any way. On the other side, the <rank> field is the most important one since it will be used as the main key to sort species and compute the final metric.
For each submitted run, please give in the submission system a description of the run. A combobox will specify wether the run was performed fully automatically or with a human assistance in the processing of the queries. Then, a textarea should contain a short description of the used method, particularly for helping differentiating the different runs submitted by the same group, for instance:
matching-based method using SIFT features, RANSAC algorithm and K-NN classifier with K=10
Optionally, you can add one or several bibtex reference(s) to publication(s) describing the method more in details.
metric
The primary metric used to evaluate the submitted runs will be a score related to the rank of the correct species in the list of retrieved species. Each test image will be attributed with a score between 0 and 1 : of 1 if the 1st returned species is correct and will decrease quickly while the rank of the correct species increases. An average score will then be computed on all test images. A simple mean on all test images would however introduce some bias. Indeed, we remind that the Pl@ntViews dataset was built in a collaborative manner. So that few contributors might have provided much more pictures than many other contributors who provided few. Since we want to evaluate the ability of a system to provide correct answers to all users, we rather measure the mean of the average classification rate per author. Furthermore, some authors sometimes provided many pictures of the same individual plant (to enrich training data with less efforts). Since we want to evaluate the ability of a system to provide the correct answer based on a single plant observation, we also have to average the classification rate on each individual plant. Finally, our primary metric is defined as the following average classification score S:
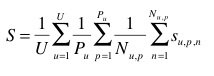
U : number of users (who have at least one image in the test data)
Pu : number of individual plants observed by the u-th user
Nu,p : number of pictures taken from the p-th plant observed by the u-th user
Su,p,n : score between 1 and 0 equals to the inverse of the rank of the correct species (for the n-th picture taken from the p-th plant observed by the u-th user)
Participants are allowed to train distinct classifiers, use different training subsets or use distinct methods for each data type.
How to register for the task
ImageCLEF has its own registration interface. Here you can choose a user name and a password. This registration interface is for example used for the submission of runs. If you already have a login from the former ImageCLEF benchmarks you can migrate it to ImageCLEF 2012 here
Schedule
- 15.12.2012: registration opens for all CLEF tasks
- 29.01.2013: training data release
- 18.03.2013: test data release
- 08.05.2013 deadline for submission of runs
- 15.05.2013: release of results
- 15.06.2013: deadline for submission of working notes
- 23-26.09.2013: CLEF 2013 Conference (Valencia)
Frequently asked questions
In the "test" dataset there are associated xml files where "type" and "content" attributes are indicated. Are we allowed to use this information during the prediction task or would it be considered as a manual intervention on the process.
- Yes, you are allowed to use this information during the prediction (like in the two previous years). We consider that species identification is a very challenging task and we don't want to add more difficulties with an organ/view prediction step.
Results
A total of 12 groups submitted 33 runs. Thanks to all participants for their efforts and their constructive feedbacks regarding the organization.
Since several participants used distinct methods for the two image categories, we give here the results in two separate tables.
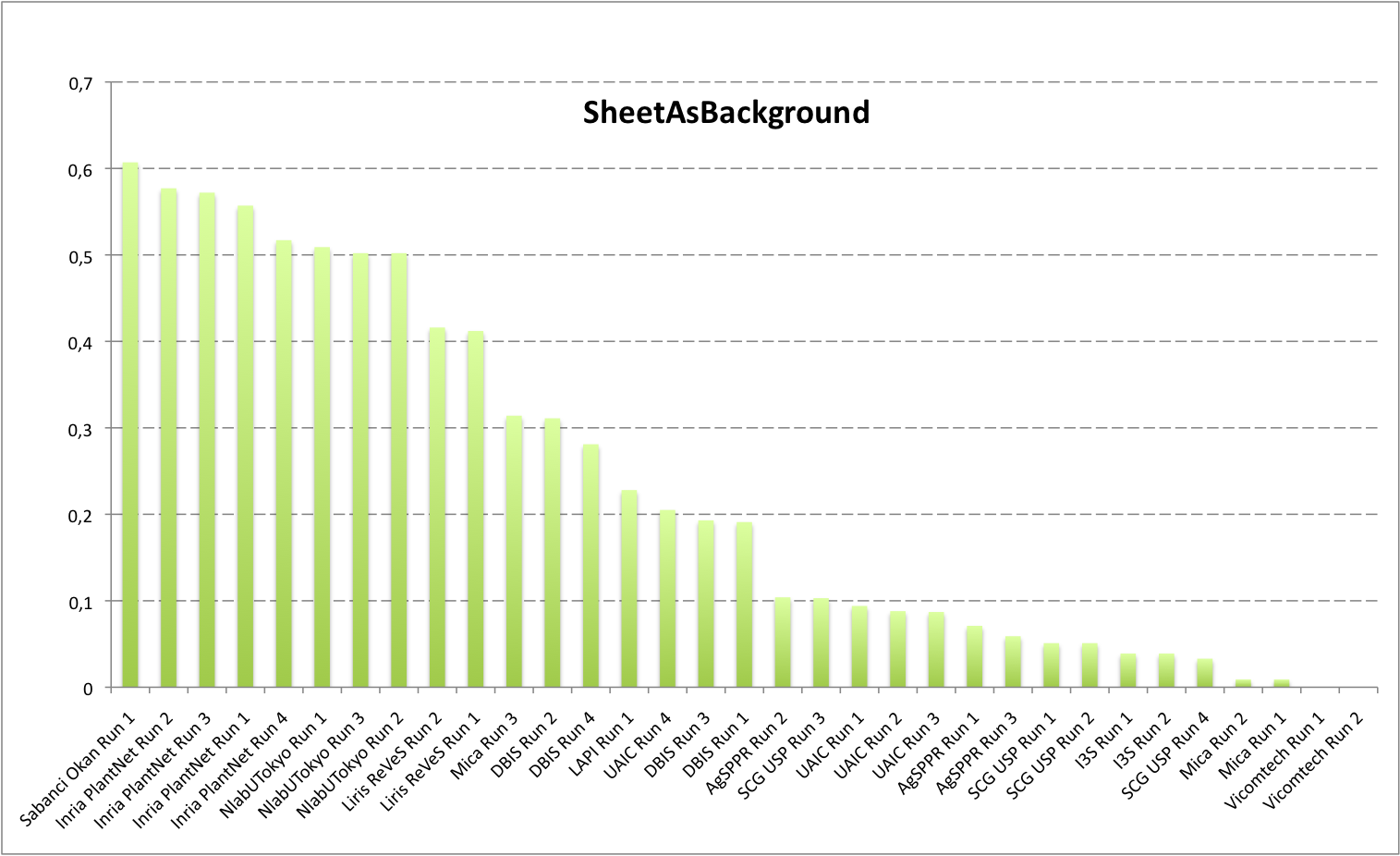
SheetAsBackground (scans and scan-like photos of single leaves)
The following table and graphic below present the scores obtained for the category "SheetAsBackground"
Click on the graphics to enlarge them.
| Run name | runfilename | retrieval type | run-type | Score | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sabanci Okan Run 1 | 1368163545166__Sabanci-Okan-Run1 | Visual | Automatic | 0,607 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Inria PlantNet Run 2 | 1367926056487__plantnet_inria_run2 | Visual | Automatic | 0,577 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Inria PlantNet Run 3 | 1367926326223__plantnet_inria_run3 | Visual | Automatic | 0,572 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Inria PlantNet Run 1 | 1367925811122__plantnet_inria_run1 | Visual | Automatic | 0,557 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Inria PlantNet Run 4 | 1368049985079__plantnet_inria_run4 | Visual | Automatic | 0,517 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| NlabUTokyo Run 1 | 1368032808839__all_siftcopphsv_cca | Visual | Automatic | 0,509 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| NlabUTokyo Run 3 | 1368041861286__run3 | Visual | Automatic | 0,502 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| NlabUTokyo Run 2 | 1368041641333__run2 | Visual | Automatic | 0,502 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Liris ReVeS Run 2 | 1367946215062__LirisReVeS_run2 | Mixed (texual + visual) | Feedback or/and human assistance | 0,416 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Liris ReVeS Run 1 | 1367946058774__LirisReVeS_run1 | Mixed (texual + visual) | Feedback or/and human assistance | 0,412 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Mica Run 3 | 1368093262111__Run3 | Visual | Automatic | 0,314 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| DBIS Run 2 | 1368038721036__DBISForMaT_run2_train2012_svm_Scan12_Photo4_-_1_4 | Visual | Automatic | 0,311 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| DBIS Run 4 | 1368045820175__DBISForMaT_run4_crossval2013_svm_feature5_config80_Photo14_1_3_3 | Visual | Automatic | 0,281 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LAPI Run 1 | 1367592085169__LAPI_run1 | Visual | Automatic | 0,228 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| UAIC Run 4 | 1368031342488__run_wiki_max_1 | Visual | Automatic | 0,205 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| DBIS Run 3 | 1368045672892__DBISForMaT_run3_crossval2013_svm_feature4_config60_1_2_3 | Visual | Automatic | 0,193 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| DBIS Run 1 | 1368038646069__DBISForMaT_run1_train2012_svm_Scan4_Photo2_1_2_3 | Visual | Automatic | 0,191 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| AgSPPR Run 2 | 1368063045390__AgSPPR_run2 | Visual | Automatic | 0,104 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SCG USP Run 3 | 1368033933190__SCG USP_run3 | Mixed (texual + visual) | Feedback or/and human assistance | 0,103 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| UAIC Run 1 | 1368028158605__run_wiki_sum_3 | Visual | Automatic | 0,094 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| UAIC Run 2 | 1368030128394__run_author10_GSP10_lire80 | Mixed (texual + visual) | Automatic | 0,088 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| UAIC Run 3 | 1368030994722__run_lire_naivebayes | Visual | Automatic | 0,087 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| AgSPPR Run 1 | 1368002237443__AgSPPR_run1 | Visual | Automatic | 0,071 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| AgSPPR Run 3 | 1368066775910__AgSPPR_run3 | Visual | Automatic | 0,059 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SCG USP Run 1 | 1367974757413__SCG USP_run1 | Mixed (texual + visual) | Automatic | 0,051 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SCG USP Run 2 | 1367975837452__SCG USP_run2 | Mixed (texual + visual) | Feedback or/and human assistance | 0,051 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| I3S Run 1 | 1368034466828__new_100 | Mixed (texual + visual) | Automatic | 0,039 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| I3S Run 2 | 1368165605197__new2_100 | Mixed (texual + visual) | Automatic | 0,039 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SCG USP Run 4 | 1368050270540__SCG USP_run4 | Mixed (texual + visual) | Feedback or/and human assistance | 0,033 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Mica Run 2 | 1366971577662__MICA-run2 | Visual | Automatic | 0,009 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Mica Run 1 | 1366945452806__MICA-run1 | Visual | Automatic | 0,009 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Vicomtech Run 1 | 1367338673606__outputCLEFTestMean | Mixed (texual + visual) | Automatic | 0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Vicomtech Run 2 | 1367338771296__outputCLEFTestMax | Mixed (texual + visual) | Automatic | 0 |
| Run name |
runfilename |
retrieval type |
run-type |
Score |
|
| NlabUTokyo Run 3 | 1368041861286__run3 | Visual | Automatic | 0,393 | |
| Inria PlantNet Run 2 | 1367926056487__plantnet_inria_run2 | Visual | Automatic | 0,385 | |
| NlabUTokyo Run 2 | 1368041641333__run2 | Visual | Automatic | 0,371 | |
| Inria PlantNet Run 1 | 1367925811122__plantnet_inria_run1 | Visual | Automatic | 0,353 | |
| NlabUTokyo Run 1 | 1368032808839__all_siftcopphsv_cca | Visual | Automatic | 0,341 | |
| Inria PlantNet Run 3 | 1367926326223__plantnet_inria_run3 | Visual | Automatic | 0,325 | |
| Inria PlantNet Run 4 | 1368049985079__plantnet_inria_run4 | Visual | Automatic | 0,245 | |
| Sabanci Okan Run 1 | 1368163545166__Sabanci-Okan-Run1 | Visual | Automatic | 0,181 | |
| DBIS Run 2 | 1368038721036__DBISForMaT_run2_train2012_svm_Scan12_Photo4_-_1_4 | Visual | Automatic | 0,159 | |
| DBIS Run 3 | 1368045672892__DBISForMaT_run3_crossval2013_svm_feature4_config60_1_2_3 | Visual | Automatic | 0,158 | |
| DBIS Run 4 | 1368045820175__DBISForMaT_run4_crossval2013_svm_feature5_config80_Photo14_1_3_3 | Visual | Automatic | 0,141 | |
| UAIC Run 4 | 1368031342488__run_wiki_max_1 | Visual | Automatic | 0,127 | |
| DBIS Run 1 | 1368038646069__DBISForMaT_run1_train2012_svm_Scan4_Photo2_1_2_3 | Visual | Automatic | 0,12 | |
| UAIC Run 1 | 1368028158605__run_wiki_sum_3 | Visual | Automatic | 0,119 | |
| UAIC Run 2 | 1368030128394__run_author10_GSP10_lire80 | Mixed (texual + visual) | Automatic | 0,117 | |
| Liris ReVeS Run 2 | 1367946215062__LirisReVeS_run2 | Mixed (texual + visual) | Feedback or/and human assistance | 0,092 | |
| Liris ReVeS Run 1 | 1367946058774__LirisReVeS_run1 | Mixed (texual + visual) | Feedback or/and human assistance | 0,089 | |
| UAIC Run 3 | 1368030994722__run_lire_naivebayes | Visual | Automatic | 0,081 | |
| Vicomtech Run 1 | 1367338673606__outputCLEFTestMean | Mixed (texual + visual) | Automatic | 0,081 | |
| Vicomtech Run 2 | 1367338771296__outputCLEFTestMax | Mixed (texual + visual) | Automatic | 0,08 | |
| LAPI Run 1 | 1367592085169__LAPI_run1 | Visual | Automatic | 0,058 | |
| Mica Run 2 | 1366971577662__MICA-run2 | Visual | Automatic | 0,053 | |
| Mica Run 3 | 1368093262111__Run3 | Visual | Automatic | 0,042 | |
| SCG USP Run 3 | 1368033933190__SCG USP_run3 | Mixed (texual + visual) | Feedback or/and human assistance | 0,03 | |
| I3S Run 1 | 1368034466828__new_100 | Mixed (texual + visual) | Automatic | 0,026 | |
| I3S Run 2 | 1368165605197__new2_100 | Mixed (texual + visual) | Automatic | 0,026 | |
| SCG USP Run 1 | 1367974757413__SCG USP_run1 | Mixed (texual + visual) | Automatic | 0,025 | |
| SCG USP Run 2 | 1367975837452__SCG USP_run2 | Mixed (texual + visual) | Feedback or/and human assistance | 0,025 | |
| Mica Run 1 | 1366945452806__MICA-run1 | Visual | Automatic | 0,023 | |
| SCG USP Run 4 | 1368050270540__SCG USP_run4 | Mixed (texual + visual) | Feedback or/and human assistance | 0,017 | |
| AgSPPR Run 2 | 1368063045390__AgSPPR_run2 | Visual | Automatic | 0 | |
| AgSPPR Run 1 | 1368002237443__AgSPPR_run1 | Visual | Automatic | 0 | |
| AgSPPR Run 3 | 1368066775910__AgSPPR_run3 | Visual | Automatic | 0 |
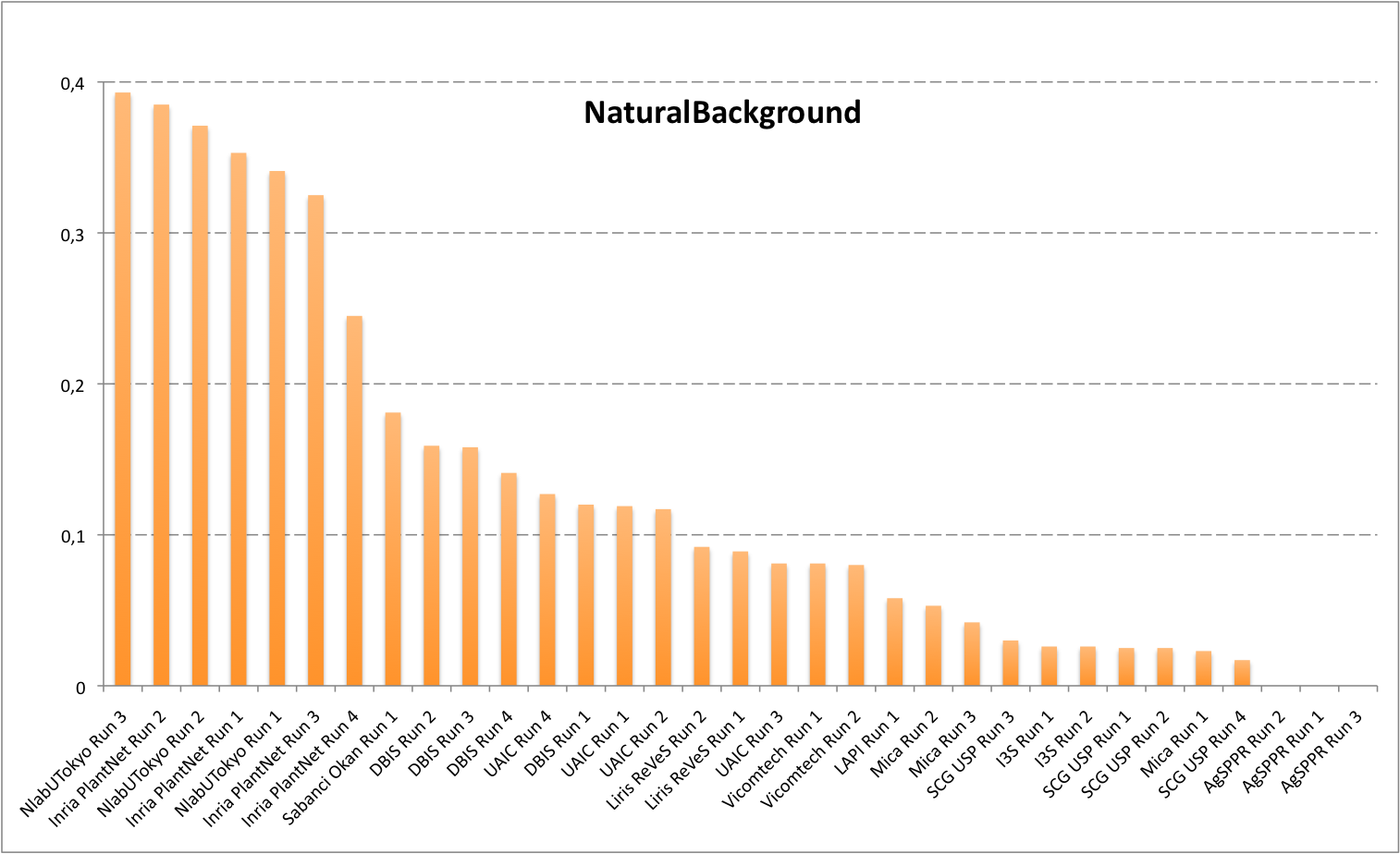
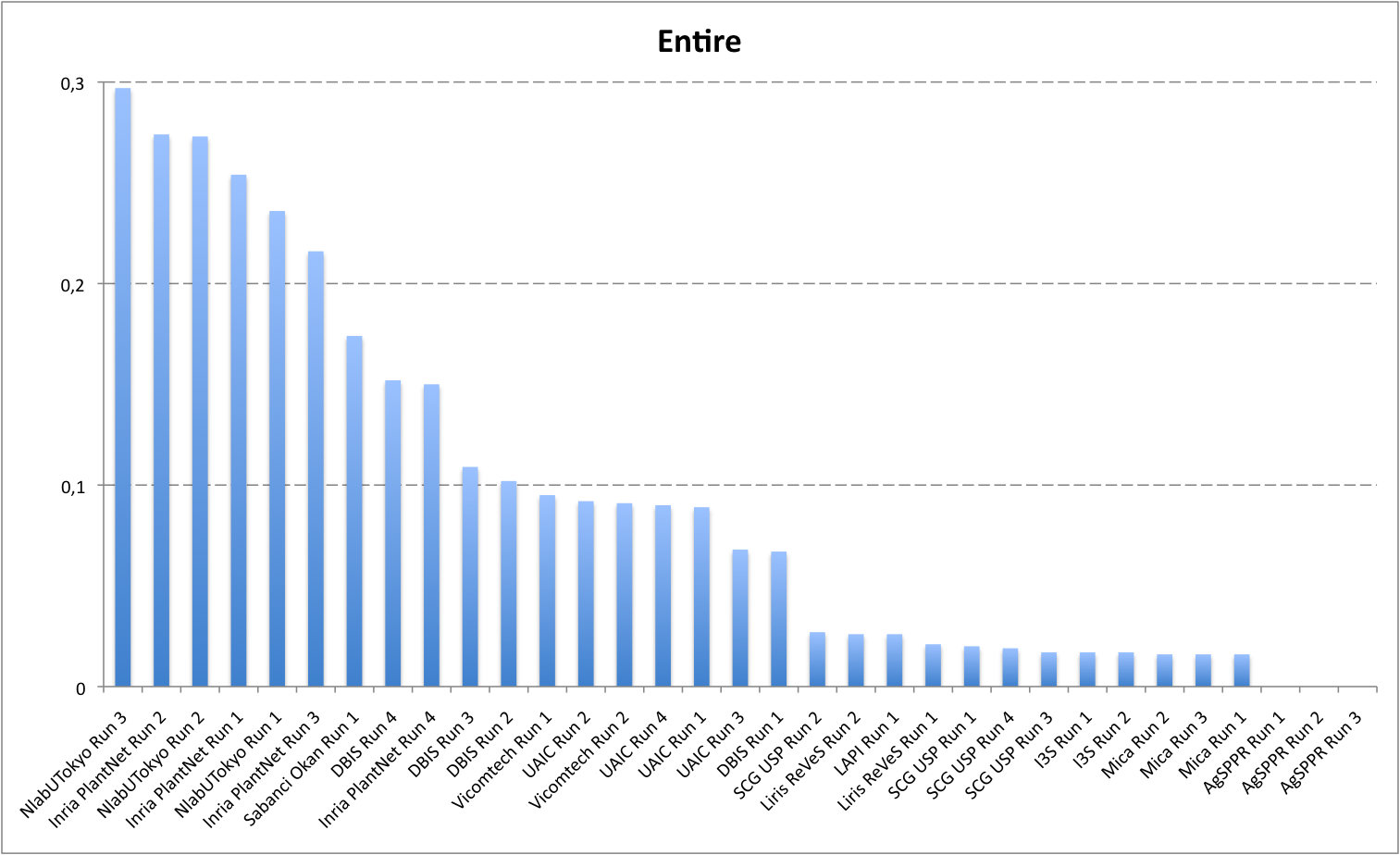
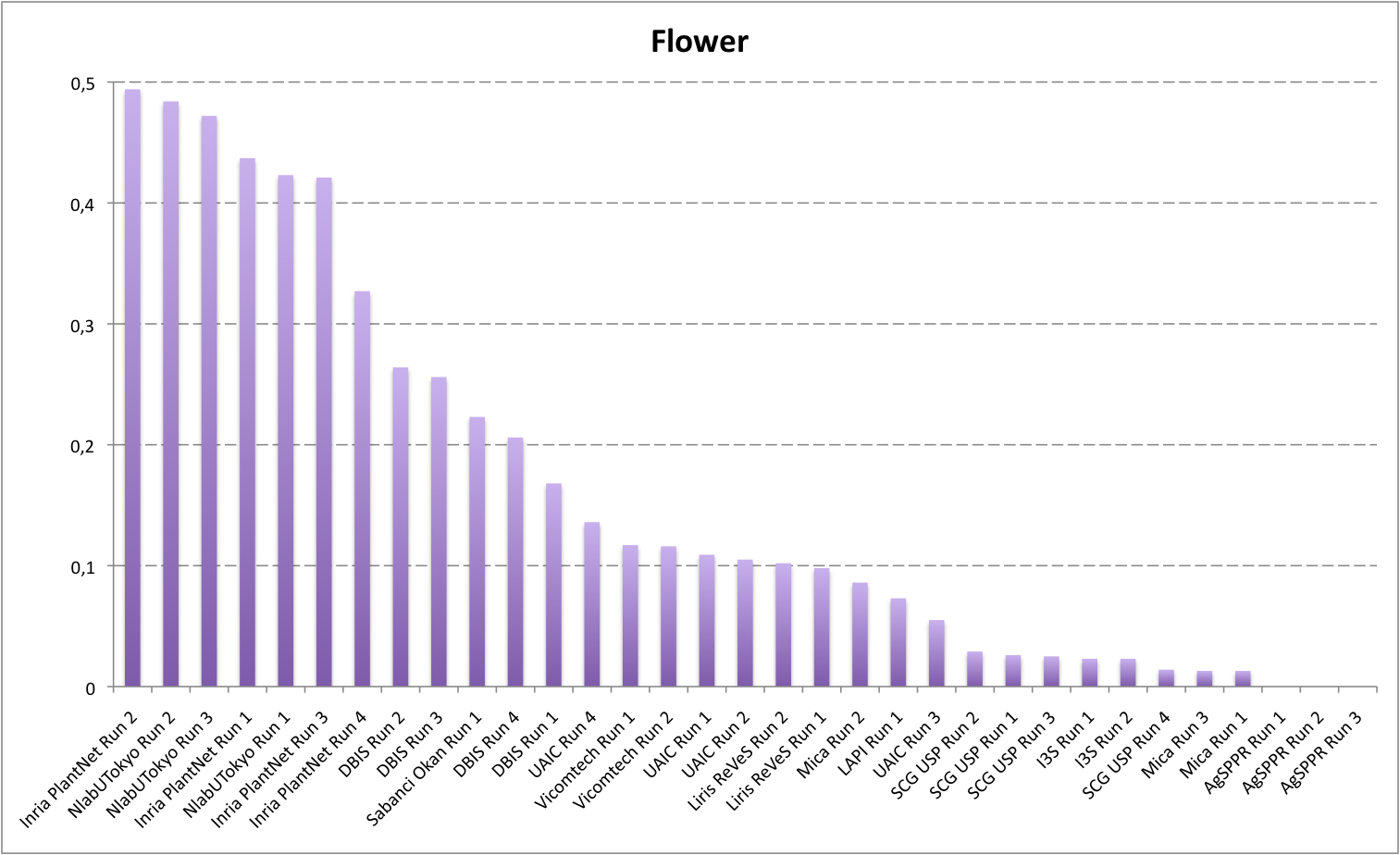
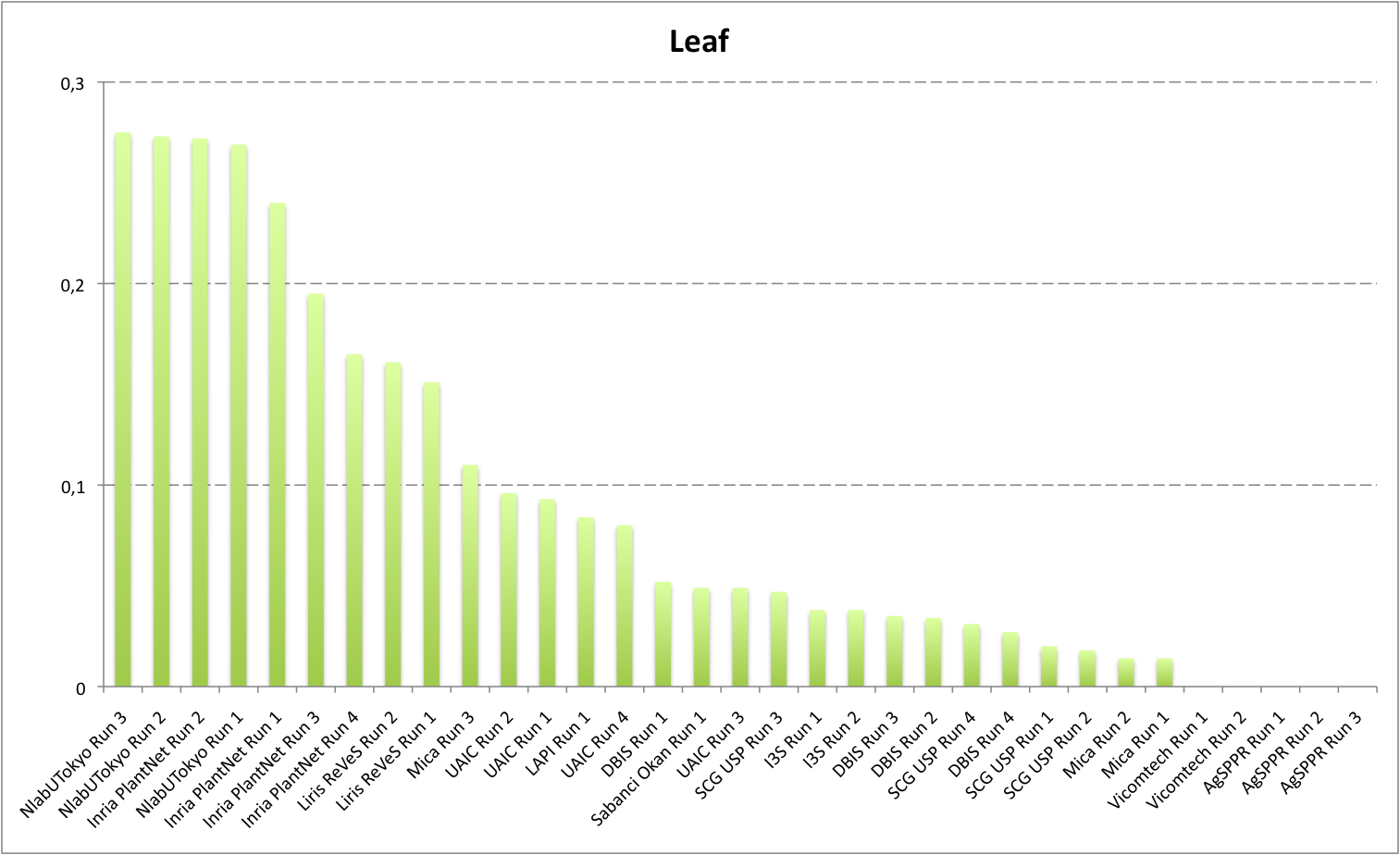
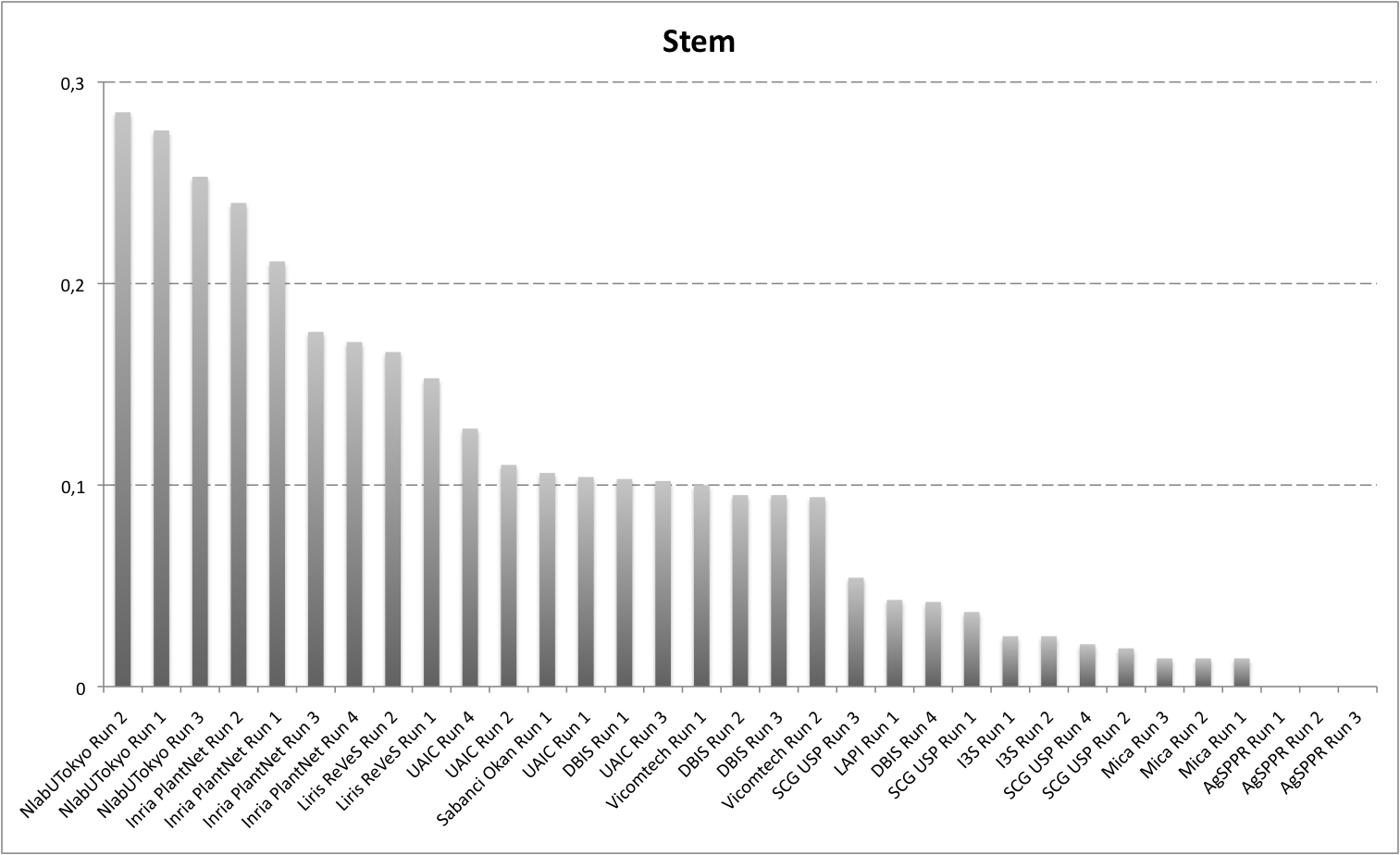
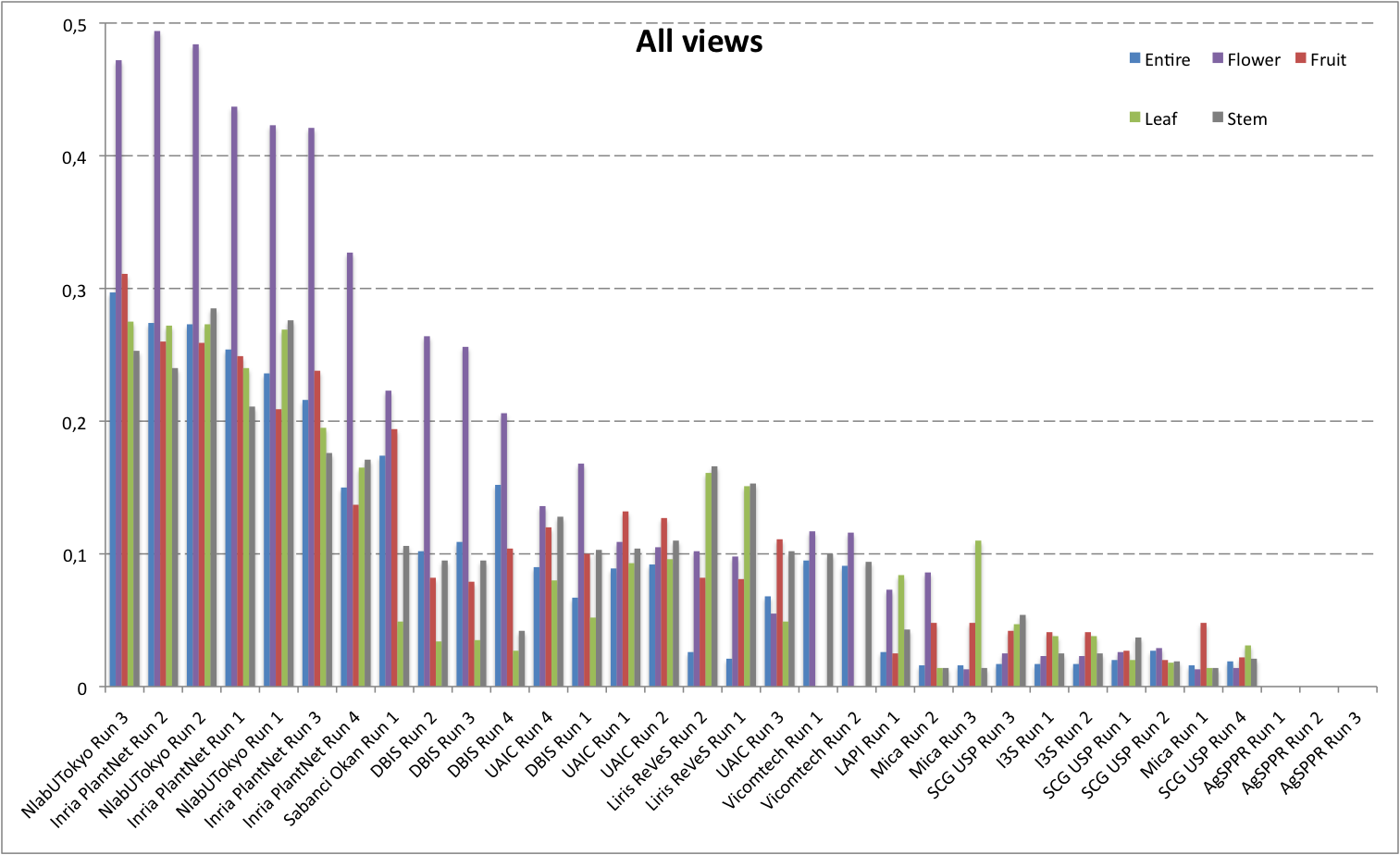
Detailed scores for each sub-categories of NaturalBackground: entire, flower, fruit, leaf, stem
The following table and graphic below present the detailed scores obtained for the sub-categories of "NaturalBackground" images. Remember that we use a specific metric weighted by authors and plants, and not by sub-categories, explaining why the NaturalBackground score is not the mean of the five scores of the sub-categories.
| Run name |
runfilename |
Entire |
Flower |
Fruit |
Leaf |
Stem |
NaturalBackground |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| NlabUTokyo Run 3 |
1368041861286__run3 |
0,297 |
0,472 |
0,311 |
0,275 |
0,253 |
0,393 |
Inria PlantNet Run 2 |
1367926056487__plantnet_inria_run2 |
0,274 |
0,494 |
0,26 |
0,272 |
0,24 |
0,385 |
NlabUTokyo Run 2 |
1368041641333__run2 |
0,273 |
0,484 |
0,259 |
0,273 |
0,285 |
0,371 |
Inria PlantNet Run 1 |
1367925811122__plantnet_inria_run1 |
0,254 |
0,437 |
0,249 |
0,24 |
0,211 |
0,353 |
NlabUTokyo Run 1 |
1368032808839__all_siftcopphsv_cca |
0,236 |
0,423 |
0,209 |
0,269 |
0,276 |
0,341 |
Inria PlantNet Run 3 |
1367926326223__plantnet_inria_run3 |
0,216 |
0,421 |
0,238 |
0,195 |
0,176 |
0,325 |
Inria PlantNet Run 4 |
1368049985079__plantnet_inria_run4 |
0,15 |
0,327 |
0,137 |
0,165 |
0,171 |
0,245 |
Sabanci Okan Run 1 |
1368163545166__Sabanci-Okan-Run1 |
0,174 |
0,223 |
0,194 |
0,049 |
0,106 |
0,181 |
DBIS Run 2 |
1368038721036__DBISForMaT_run2_train2012_svm_Scan12_Photo4_-_1_4 |
0,102 |
0,264 |
0,082 |
0,034 |
0,095 |
0,159 |
DBIS Run 3 |
1368045672892__DBISForMaT_run3_crossval2013_svm_feature4_config60_1_2_3 |
0,109 |
0,256 |
0,079 |
0,035 |
0,095 |
0,158 |
DBIS Run 4 |
1368045820175__DBISForMaT_run4_crossval2013_svm_feature5_config80_Photo14_1_3_3 |
0,152 |
0,206 |
0,104 |
0,027 |
0,042 |
0,141 |
UAIC Run 4 |
1368031342488__run_wiki_max_1 |
0,09 |
0,136 |
0,12 |
0,08 |
0,128 |
0,127 |
DBIS Run 1 |
1368038646069__DBISForMaT_run1_train2012_svm_Scan4_Photo2_1_2_3 |
0,067 |
0,168 |
0,1 |
0,052 |
0,103 |
0,12 |
UAIC Run 1 |
1368028158605__run_wiki_sum_3 |
0,089 |
0,109 |
0,132 |
0,093 |
0,104 |
0,119 |
UAIC Run 2 |
1368030128394__run_author10_GSP10_lire80 |
0,092 |
0,105 |
0,127 |
0,096 |
0,11 |
0,117 |
Liris ReVeS Run 2 |
1367946215062__LirisReVeS_run2 |
0,026 |
0,102 |
0,082 |
0,161 |
0,166 |
0,092 |
Liris ReVeS Run 1 |
1367946058774__LirisReVeS_run1 |
0,021 |
0,098 |
0,081 |
0,151 |
0,153 |
0,089 |
UAIC Run 3 |
1368030994722__run_lire_naivebayes |
0,068 |
0,055 |
0,111 |
0,049 |
0,102 |
0,081 |
Vicomtech Run 1 |
1367338673606__outputCLEFTestMean |
0,095 |
0,117 |
0 |
0 |
0,1 |
0,081 |
Vicomtech Run 2 |
1367338771296__outputCLEFTestMax |
0,091 |
0,116 |
0 |
0 |
0,094 |
0,08 |
LAPI Run 1 |
1367592085169__LAPI_run1 |
0,026 |
0,073 |
0,025 |
0,084 |
0,043 |
0,058 |
Mica Run 2 |
1366971577662__MICA-run2 |
0,016 |
0,086 |
0,048 |
0,014 |
0,014 |
0,053 |
Mica Run 3 |
1368093262111__Run3 |
0,016 |
0,013 |
0,048 |
0,11 |
0,014 |
0,042 |
SCG USP Run 3 |
1368033933190__SCG USP_run3 |
0,017 |
0,025 |
0,042 |
0,047 |
0,054 |
0,03 |
I3S Run 1 |
1368034466828__new_100 |
0,017 |
0,023 |
0,041 |
0,038 |
0,025 |
0,026 |
I3S Run 2 |
1368165605197__new2_100 |
0,017 |
0,023 |
0,041 |
0,038 |
0,025 |
0,026 |
SCG USP Run 1 |
1367974757413__SCG USP_run1 |
0,02 |
0,026 |
0,027 |
0,02 |
0,037 |
0,025 |
SCG USP Run 2 |
1367975837452__SCG USP_run2 |
0,027 |
0,029 |
0,02 |
0,018 |
0,019 |
0,025 |
Mica Run 1 |
1366945452806__MICA-run1 |
0,016 |
0,013 |
0,048 |
0,014 |
0,014 |
0,023 |
SCG USP Run 4 |
1368050270540__SCG USP_run4 |
0,019 |
0,014 |
0,022 |
0,031 |
0,021 |
0,017 |
AgSPPR Run 1 |
1368002237443__AgSPPR_run1 |
0 |
0 |
0 |
0 |
0 |
0 |
AgSPPR Run 2 |
1368063045390__AgSPPR_run2 |
0 |
0 |
0 |
0 |
0 |
0 |
AgSPPR Run 3 |
1368066775910__AgSPPR_run3 |
0 |
0 |
0 |
0 |
0 |
0 |
|
Click on the graphics to enlarge them.
Additional statistics and comments on these results will be provided in the overview working note of the task published within CLEF 2013.
_____________________________________________________________________________________________________________________
Contacts
Hervé Goeau (INRIA-ZENITH, INRIA-IMEDIA): herve(replace-that-by-a-dot)goeau(replace-that-by-an-arrobe)inria.fr
Alexis Joly (INRIA-ZENITH): alexis(replace-that-by-a-dot)joly(replace-that-by-an-arrobe)inria.fr
Pierre Bonnet (AMAP): pierre(replace-that-by-a-dot)bonnet(replace-that-by-an-arrobe)cirad.fr
| Attachment | Size |
|---|---|
| 142.66 KB | |
| 376.74 KB | |
| 311.49 KB | |
| 1.37 MB | |
| 349.82 KB | |
| 1.01 MB | |
| 283.51 KB | |
| 245.24 KB | |
| 1.02 KB | |
| 979 bytes | |
| 4.51 MB | |
| 192.77 KB | |
| 160.67 KB | |
| 169.68 KB | |
| 182.3 KB | |
| 154.76 KB | |
| 166.65 KB | |
| 131.41 KB | |
| 122.79 KB |